β-lactamases (bla TEM, bla SHV, bla CTXM-1, bla VEB, bla OXA-1 ) and class C β-lactamases gene frequency in Pseudomonas aeruginosa isolated from various clinical specimens in Khartoum State, Sudan: a cross sectional study
- PMID: 33363717
- PMCID: PMC7737708
- DOI: 10.12688/f1000research.24818.3
β-lactamases (bla TEM, bla SHV, bla CTXM-1, bla VEB, bla OXA-1 ) and class C β-lactamases gene frequency in Pseudomonas aeruginosa isolated from various clinical specimens in Khartoum State, Sudan: a cross sectional study
Abstract
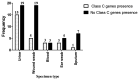
Background:Pseudomonas aeruginosa is a pathogenic bacterium, causing nosocomial infections with intrinsic and acquired resistance mechanisms to a large group of antibiotics, including β-lactams. This study aimed to determine the susceptibility pattern to selected antibiotics and to index the first reported β-lactamases genes frequency in Ps. aeruginosa in Khartoum State, Sudan. Methods: 121 Ps. aeruginosa clinical isolates from various clinical specimens were used in this cross sectional study conducted in Khartoum State. Eighty isolates were confirmed as Ps.aeruginosa through conventional identification methods and species specific primers. The susceptibility pattern of the confirmed isolates to selected antibiotics was done following the Kirby Bauer disk diffusion method. Multiplex PCR was used for detection of seven β-lactamase genes ( blaTEM, blaSHV, blaCTXM-1, blaVEB, blaOXA-1, blaAmpC and blaDHA). Results: Of the 80 confirmed Ps. aeruginosa isolates, 8 (10%) were resistant to Imipenem while all isolates were resistant to Amoxicillin and Amoxyclav (100%). A total of 43 (54%) Ps. aeruginosa isolates were positive for blaTEM, blaSHV, blaCTXM-1, blaVEB and blaOXA-1 genes, while 27 (34%) were positive for class C β- Lactamases, and 20 (25%) were positive for both classes. Frequency of beta-lactamases genes was as follows: blaTEM, 19 (44.2%); blaSHV, 16 (37.2%); bla CTX-M1, 10 (23.3%); blaVEB, 14 (32.6%); blaOXA-1, 7 (16.3%). blaAmpC 22 (81.5%) and bla DHA 8 (29.6%). In total, 3 (11.1%) isolates were positive for both bla AmpC and blaDHA genes. Conclusion:Ps. aeruginosa isolates showed a high rate of β- lactamases production, with co-resistance to other antibiotic classes. The lowest resistance rate of Ps. aeruginosa was to Imipenem followed by Gentamicin and Ciprofloxacin. No statistically significant relationship between production of β-lactamases in Ps. aeruginosa and resistance to third generation cephalosporins was found.
Keywords: ESBLs; Khartoum-Sudan.; Polymerase Chain Reaction; Ps. aeruginosa; class C β-lactamase; pyocyanin pigment.
Copyright: © 2021 Abdelrahman DN et al.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
-
- Forbes BA, Sahm DF, Weissfeld AS: Study guide for Bailey & Scott’s diagnostic microbiology. Mosby, USA;2007. Reference Source
-
- Agouri SR: Genetic characterisation of MBL positive pseudomonas and Enterobacteriaceae. Cardiff University;2014. Reference Source
-
- Beta-lactamases B, d Inhibitor Resistant T : Extended spectrum beta-lactamases.
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

