Discovery of Novel Biosynthetic Gene Cluster Diversity From a Soil Metagenomic Library
- PMID: 33365020
- PMCID: PMC7750434
- DOI: 10.3389/fmicb.2020.585398
Discovery of Novel Biosynthetic Gene Cluster Diversity From a Soil Metagenomic Library
Abstract
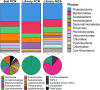
Soil microorganisms historically have been a rich resource for natural product discovery, yet the majority of these microbes remain uncultivated and their biosynthetic capacity is left underexplored. To identify the biosynthetic potential of soil microorganisms using a culture-independent approach, we constructed a large-insert metagenomic library in Escherichia coli from a topsoil sampled from the Cullars Rotation (Auburn, AL, United States), a long-term crop rotation experiment. Library clones were screened for biosynthetic gene clusters (BGCs) using either PCR or a NGS (next generation sequencing) multiplexed pooling strategy, coupled with bioinformatic analysis to identify contigs associated with each metagenomic clone. A total of 1,015 BGCs were detected from 19,200 clones, identifying 223 clones (1.2%) that carry a polyketide synthase (PKS) and/or a non-ribosomal peptide synthetase (NRPS) cluster, a dramatically improved hit rate compared to PCR screening that targeted type I polyketide ketosynthase (KS) domains. The NRPS and PKS clusters identified by NGS were distinct from known BGCs in the MIBiG database or those PKS clusters identified by PCR. Likewise, 16S rRNA gene sequences obtained by NGS of the library included many representatives that were not recovered by PCR, in concordance with the same bias observed in KS amplicon screening. This study provides novel resources for natural product discovery and circumvents amplification bias to allow annotation of a soil metagenomic library for a more complete picture of its functional and phylogenetic diversity.
Keywords: biases; biosynthetic ability; metagenome; next-generating sequencing; soil.
Copyright © 2020 Santana-Pereira, Sandoval-Powers, Monsma, Zhou, Santos, Mead and Liles.
Conflict of interest statement
ML and DM are the cofounders of the Varigen Biosciences Corporation. A licensing agreement between Auburn University and the Varigen Biosciences Corporation has been established for commercial development of the Cullars soil metagenomic library described in this manuscript. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

