Current Methods for Detecting Cell Membrane Transient Interactions
- PMID: 33365301
- PMCID: PMC7750205
- DOI: 10.3389/fchem.2020.603259
Current Methods for Detecting Cell Membrane Transient Interactions
Abstract
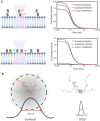
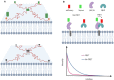
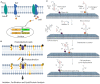
Short-lived cell membrane complexes play a key role in regulating cell signaling and communication. Many of these complexes are formed based on low-affinity and transient interactions among various lipids and proteins. New techniques have emerged to study these previously overlooked membrane transient interactions. Exciting functions of these transient interactions have been discovered in cellular events such as immune signaling, host-pathogen interactions, and diseases such as cancer. In this review, we have summarized current experimental methods that allow us to detect and analyze short-lived cell membrane protein-protein, lipid-protein, and lipid-lipid interactions. These methods can provide useful information about the strengths, kinetics, and/or spatial patterns of membrane transient interactions. However, each method also has its own limitations. We hope this review can be used as a guideline to help the audience to choose proper approaches for studying membrane transient interactions in different membrane trafficking and cell signaling events.
Keywords: cell signaling; lipids; membrane biology; membrane probes; proteins; transient interactions.
Copyright © 2020 Bagheri, Ali and You.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

