Survey and Sequence Characterization of Bovine Mastitis-Associated Escherichia coli in Dairy Herds
- PMID: 33365333
- PMCID: PMC7750360
- DOI: 10.3389/fvets.2020.582297
Survey and Sequence Characterization of Bovine Mastitis-Associated Escherichia coli in Dairy Herds
Abstract
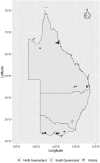
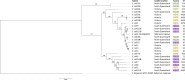
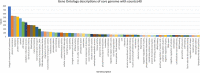
Escherichia coli is frequently associated with mastitis in cattle. "Pathogenic" and "commensal" isolates appear to be genetically similar. With a few exceptions, no notable genotypic differences have been found between commensal and mastitis-associated E. coli. In this study, 24 E. coli strains were isolated from dairy cows with clinical mastitis in three geographic regions of Australia (North Queensland, South Queensland, and Victoria), sequenced, then genomically surveyed. There was no observed relationship between sequence type (ST) and region (p = 0.51). The most common Multi Locus Sequence Type was ST10 (38%), then ST4429 (13%). Pangenomic analysis revealed a soft-core genome of 3,463 genes, including genes associated with antibiotic resistance, chemotaxis, motility, adhesion, biofilm formation, and pili. A total of 36 different plasmids were identified and generally found to have local distributions (p = 0.02). Only 2 plasmids contained antibiotic resistance genes, a p1303_5-like plasmid encoding multidrug-resistance (trimethoprim, quaternary ammonium, beta-lactam, streptomycin, sulfonamide, and kanamycin) from two North Queensland isolates on the same farm, while three Victorian isolates from the same farm contained a pCFSAN004177P_01-like plasmid encoding tetracycline-resistance. This pattern is consistent with a local spread of antibiotic resistance through plasmids of bovine mastitis cases. Notably, co-occurrence of plasmids containing virulence factors/antibiotic resistance with putative mobilization was rare, though the multidrug resistant p1303_5-like plasmid was predicted to be conjugative and is of some concern. This survey has provided greater understanding of antibiotic resistance within E. coli-associated bovine mastitis which will allow greater prediction and improved decision making in disease management.
Keywords: antibiotic resistance; cattle; comparative genomics; genomics; plasmid; virulence; whole genome sequencing.
Copyright © 2020 Alawneh, Vezina, Ramay, Al-Harbi, James, Soust, Moore and Olchowy.
Conflict of interest statement
MS was employed by the company Terragen Biotech Pty Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

