Characterization of the complete chloroplast genome sequence of Galinsoga parviflora and its phylogenetic implications
- PMID: 33365429
- PMCID: PMC7687395
- DOI: 10.1080/23802359.2019.1623106
Characterization of the complete chloroplast genome sequence of Galinsoga parviflora and its phylogenetic implications
Abstract
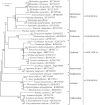
Galinsoga parviflora is an invasive weed in southwest of Chinese agricultural systems and commonly used as medicine and food. In this study, the complete chloroplast genome of the G. parviflora was assembled from the whole genome Illumina sequencing data. The circular genome is 151,811 bp in size, which composed of one large single-copy (LSC) and one small single-copy (SSC) regions of 83,594 bp and 18,141 bp, respectively, and separated by a pair of inverted repeat (IR) regions of 25,038 bp each. It encodes a total of 113 gene species (80 protein-coding, 29 tRNA, and four rRNA species), in which 19 of them with double copies. The overall GC content is 37.7% while the GC content of the LSC, SSC, and IR regions are 35.8%, 31.3%, and 43.1%, separately. Phylogenetic analysis indicated that Galinsoga parviflora was closely related to Galinsoga quadriradiata.
Keywords: Galinsoga parviflora; chloroplast genome; phylogenetic analysis; weed.
© 2019 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Figures
Similar articles
-
Characterization of the complete chloroplast genome sequence of Fritillaria delavayi, an ethnomedicinal plant in China.Mitochondrial DNA B Resour. 2019 Oct 10;4(2):3492-3493. doi: 10.1080/23802359.2019.1674742. Mitochondrial DNA B Resour. 2019. PMID: 33366054 Free PMC article.
-
Chloroplast genome of Gaura parviflora Douglas and its comparative analysis.Mitochondrial DNA B Resour. 2021 Mar 11;6(3):760-761. doi: 10.1080/23802359.2021.1878960. Mitochondrial DNA B Resour. 2021. PMID: 33763571 Free PMC article.
-
Complete chloroplast genome sequence of Rosa roxburghii and its phylogenetic analysis.Mitochondrial DNA B Resour. 2018 Feb 1;3(1):149-150. doi: 10.1080/23802359.2018.1431074. Mitochondrial DNA B Resour. 2018. PMID: 33490491 Free PMC article.
-
The Complete Chloroplast Genome of Endangered Species Stemona parviflora: Insight into the Phylogenetic Relationship and Conservation Implications.Genes (Basel). 2022 Jul 29;13(8):1361. doi: 10.3390/genes13081361. Genes (Basel). 2022. PMID: 36011272 Free PMC article.
-
Characterization of the complete chloroplast genome of Festuca sinensis (Gramineae).Mitochondrial DNA B Resour. 2019 Dec 27;5(1):358-359. doi: 10.1080/23802359.2019.1704192. Mitochondrial DNA B Resour. 2019. PMID: 33366555 Free PMC article.
Cited by
-
Galinsoga parviflora (Cav.): A comprehensive review on ethnomedicinal, phytochemical and pharmacological studies.Heliyon. 2023 Feb 8;9(2):e13517. doi: 10.1016/j.heliyon.2023.e13517. eCollection 2023 Feb. Heliyon. 2023. PMID: 36846665 Free PMC article. Review.
-
The complete chloroplast genome sequence of Elsholtzia densa, a herb with volatile aroma component.Mitochondrial DNA B Resour. 2020 Jan 14;5(1):595-596. doi: 10.1080/23802359.2019.1710597. Mitochondrial DNA B Resour. 2020. PMID: 33366662 Free PMC article.
-
The complete chloroplast genome of Leibnitzia nepalensis (Kunze) Kitamura, 1983 (Asteraceae, Mutisieae) and its phylogenetic analysis.Mitochondrial DNA B Resour. 2025 Feb 11;10(3):212-217. doi: 10.1080/23802359.2025.2463501. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 39944031 Free PMC article.
-
The complete chloroplast genome of Gaillardia pulchella Foug. and its phylogenetic analysis.Mitochondrial DNA B Resour. 2025 Aug 23;10(9):863-867. doi: 10.1080/23802359.2025.2550611. eCollection 2025. Mitochondrial DNA B Resour. 2025. PMID: 40861888 Free PMC article.
References
-
- Ali S, Zameer S, Yaqoob M. 2017. Ethnobotanical, phytochemical and pharmacological properties of Galinsoga parviflora (Asteraceae): a review. Trop J Pharm Res. 12:3023–3033.
-
- Bazylko A, Stolarczyk M, Derwińska M, et al. . 2012. Determination of antioxidant activity of extracts and fractions obtained from Galinsoga parviflora and Galinsoga quadriradiata, and a qualitative study of the most active fractions using TLC and HPLC methods. Nat Prod Res. 17:1584–1593. - PubMed
-
- Bazylko A, Borzym J, Parzonko A. 2015. Determination of in vitro antioxidant and UV-protecting activity of aqueous and ethanolic extracts from Galinsoga parviflora and Galinsoga quadriradiata herb. J Photochem Photobiol B Biol. 149:189–195. - PubMed
-
- Chipurura B, Muchuweti M, Parawira W, et al. . 2009. An assessment of the phenolic content, composition and antioxidant capacity of Bidens pilosa, Cleome gynandra, Corchorus olitorius, Galinsoga parviflora and Amaranthus hybridus. I All Africa Horticultural Congress 911; p. 417–426.
-
- Damalas CA. 2008. Distribution, biology, and agricultural importance of Galinsoga parviflora (Asteraceae). Weed Biol Manag. 3:147–153.
LinkOut - more resources
Full Text Sources
Miscellaneous
