Characterization of the complete chloroplast genome sequence of Fritillaria delavayi, an ethnomedicinal plant in China
- PMID: 33366054
- PMCID: PMC7707188
- DOI: 10.1080/23802359.2019.1674742
Characterization of the complete chloroplast genome sequence of Fritillaria delavayi, an ethnomedicinal plant in China
Abstract
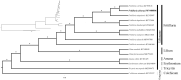
Fritillaria delavayi has been widely used as a traditional Chinese medicine (TCM) to treat respiratory diseases for thousands of years. In this study, the complete chloroplast genome of F. delavayi was assembled. The circular genome is 151,938 bp in size, which is comprised of one large single-copy (LSC) region of 81,757 bp and one small single-copy (SSC) region of 17,537 bp and separated by a pair of inverted repeat (IR) regions of 26,322 bp. A total of 112 unique genes (78 protein-coding, 30 tRNA, and 4 rRNA) are predicted and 19 of them are duplicated in IR regions. The overall GC content is 37.0% while the GC content of the LSC, SSC, and IR regions are 34.8, 30.5, and 42.5%, separately. Phylogenetic analysis indicated that F. delavayi was closely related to F. cirrhosa.
Keywords: Fritillaria delavayi; chloroplast genome; phylogenetic analysis.
© 2019 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
Similar articles
-
The complete chloroplast genome sequence of the medicinal plant Paeonia delavayi Franchet. (Paeoniaceae).Mitochondrial DNA B Resour. 2019 Oct 15;4(2):3553-3555. doi: 10.1080/23802359.2019.1677528. Mitochondrial DNA B Resour. 2019. PMID: 33366082 Free PMC article.
-
The complete chloroplast genome of Magnolia delavayi, a threatened species endemic to Southwest China.Mitochondrial DNA B Resour. 2020 Jul 16;5(3):2734-2735. doi: 10.1080/23802359.2019.1660252. Mitochondrial DNA B Resour. 2020. PMID: 33457926 Free PMC article.
-
Characterisation of the complete chloroplast genome of Gentiana delavayi Franch. (gentianaceae), a medicinal plant in southwest of China.Mitochondrial DNA B Resour. 2019 Oct 18;4(2):3638-3639. doi: 10.1080/23802359.2019.1677196. Mitochondrial DNA B Resour. 2019. PMID: 33366120 Free PMC article.
-
Characterization of the complete chloroplast genome sequence of Galinsoga parviflora and its phylogenetic implications.Mitochondrial DNA B Resour. 2019 Jul 10;4(2):2106-2108. doi: 10.1080/23802359.2019.1623106. Mitochondrial DNA B Resour. 2019. PMID: 33365429 Free PMC article.
-
The Complete Chloroplast Genome Sequences of Fritillaria ussuriensis Maxim. and Fritillaria cirrhosa D. Don, and Comparative Analysis with Other Fritillaria Species.Molecules. 2017 Jun 13;22(6):982. doi: 10.3390/molecules22060982. Molecules. 2017. PMID: 28608849 Free PMC article.
References
-
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 27:578–579. - PubMed
-
- Li Q, Li Y, Song J, Xu H, Xu J, Zhu Y, Li X, Gao H, Dong L, Qian J. 2014. High-accuracy de novo assembly and SNP detection of chloroplast genomes using a SMRT circular consensus sequencing strategy. New Phytol. 204:1041–1049. - PubMed
-
- Li S-L, Chan S-W, Li P, Lin G, Zhou G-h, Ren Y-J, Chiu FC-K. 1999. Pre-column derivatization and gas chromatographic determination of alkaloids in bulbs of Fritillaria. J Chromatogr A. 859:183–192. - PubMed
-
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). 2010:1–8. IEEE.
LinkOut - more resources
Full Text Sources
Miscellaneous
