The complete chloroplast genome sequence of Styrax odoratissimus (Styracaceae)
- PMID: 33366313
- PMCID: PMC7707729
- DOI: 10.1080/23802359.2019.1688705
The complete chloroplast genome sequence of Styrax odoratissimus (Styracaceae)
Abstract
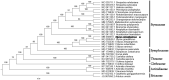
Styrax odoratissimus Champ. ex Benth., also known as the extraordinary aromatic plant of Styracaceae, has been seriously threatened due to human activities. In this study, the complete chloroplast (cp) genome sequence of S. odoratissimus was determined by applying next-generation sequencing. The entire cp genome was determined to be 157,921 bp in length. It contained large single-copy (LSC) and small single-copy (SSC) regions of 87,524 bp and 18,301 bp, respectively, which were separated by a pair of 26,048 bp inverted repeat (IR) regions. The genome contained 130 genes, including 85 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The overall GC content of S. odoratissimus genome is 36.96% and the corresponding values in LSC, SSC, and IR regions are 34.80, 30.27, and 42.92%, respectively. A phylogenetic tree reconstructed by 31 chloroplast genomes reveals that S. odoratissimus is most related to Styrax grandiflorus Griffith.
Keywords: Styracaceae; Styrax odoratissimus; complete chloroplast genome; phylogenetic analysis.
© 2019 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
References
-
- Huang S-M, James WG. 2003. Styracaceae In: Wu Z-Y, Raven PH, Hong D-Y, editors. Flora of China (Styracaceae). Vol. 15 Beijing: Science Press/St. Louis: Missouri Botanic Garden Press; p. 258.
-
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. . 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374. - PMC - PubMed
-
- Katoh K, Standley DM. 2014. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 1079:131–146. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous