The complete mitochondrial genome of Acrossocheilus yunnanensis (Teleostei: Cypriniformes: Cyprinidae) and its phylogenetic position
- PMID: 33366318
- PMCID: PMC7707677
- DOI: 10.1080/23802359.2019.1688100
The complete mitochondrial genome of Acrossocheilus yunnanensis (Teleostei: Cypriniformes: Cyprinidae) and its phylogenetic position
Abstract
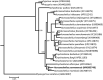
The complete mitochondrial genome of Acrossocheilus yunnanensis was determined in this study. It contained 13 protein-coding genes (PCGs), 22 tRNA, 2 rRNAs, and a control region with the base composition 31.47% A, 27.83% C, 24.65% T, and 16.05% G. Here we compared this newly determined mitogenome with another one from the same species reported before. The variable sites and the genetic distances between the two mitogenomes were 134 bp and 0.8%. Sixty-five variable sites occurred in the PCGs. The results from the phylogenetic analysis showed that the genus Acrossocheilus is not a monophyletic group and Acrossocheilus yunnanensis demonstrates a close relationship with Acrossocheilus monticola.
Keywords: Acrossocheilus yunnanensis; Cyprinidae; Mitochondrial DNA; phylogeny.
© 2019 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
References
-
- Froese R, Pauly D, editors. 2019. FishBase. World Wide Web electronic publication; [accessed 2019 September 2]. http://www.fishbase.org, version (08/2019).
-
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818. - PubMed
LinkOut - more resources
Full Text Sources