The complete chloroplast genome of Vatica guangxiensis S.L. Mo (Dipterocarpaceae): genome structure and evolution
- PMID: 33367086
- PMCID: PMC7671599
- DOI: 10.1080/23802359.2020.1835580
The complete chloroplast genome of Vatica guangxiensis S.L. Mo (Dipterocarpaceae): genome structure and evolution
Abstract
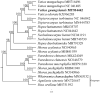
Vatica guangxiensis S.L. Mo is an evergreen large tree of Dipterocarpaceae. Herein, we assembled the complete chloroplast genome of Vatica guangxiensis by next-generation sequencing technologies. The complete chloroplast genome sequence of Vatica guangxiensis is 151,010 base pairs (bp) in length, including a pair of inverted repeat regions (IRs, 23,827 bp), one large single-copy region (LSC, 83,353 bp), one small single-copy region (SSC, 20,003 bp). Besides, the complete chloroplast genome contains 123 genes in total, including 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that Vatica guangxiensis has the closest relationship with Vatica mangachapoi. Our study lay a foundation for further research of Vatica mangachapoi.
Keywords: Chloroplast genome; Dipterocarpaceae; Vatica guangxiensis; phylogenetic analysis.
© 2020 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures
References
-
- Bi G, Mao Y, Xing Q, Cao M.. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22. - PubMed
-
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sym-Posium Series. 41:95–98.
-
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ.. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv, 256479
LinkOut - more resources
Full Text Sources