SnRK1 Phosphorylates and Destabilizes WRKY3 to Enhance Barley Immunity to Powdery Mildew
- PMID: 33367247
- PMCID: PMC7747994
- DOI: 10.1016/j.xplc.2020.100083
SnRK1 Phosphorylates and Destabilizes WRKY3 to Enhance Barley Immunity to Powdery Mildew
Abstract
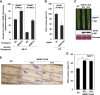
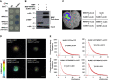
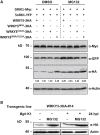
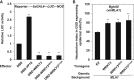
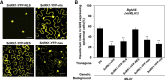
Plants recognize pathogens and activate immune responses, which usually involve massive transcriptional reprogramming. The evolutionarily conserved kinase, Sucrose non-fermenting-related kinase 1 (SnRK1), functions as a metabolic regulator that is essential for plant growth and stress responses. Here, we identify barley SnRK1 and a WRKY3 transcription factor by screening a cDNA library. SnRK1 interacts with WRKY3 in yeast, as confirmed by pull-down and luciferase complementation assays. Förster resonance energy transfer combined with noninvasive fluorescence lifetime imaging analysis indicates that the interaction occurs in the barley nucleus. Transient expression and virus-induced gene silencing analyses indicate that WRKY3 acts as a repressor of disease resistance to the Bgh fungus. Barley plants overexpressing WRKY3 have enhanced fungal microcolony formation and sporulation. Phosphorylation assays show that SnRK1 phosphorylates WRKY3 mainly at Ser83 and Ser112 to destabilize the repressor, and WRKY3 non-phosphorylation-null mutants at these two sites are more stable than the wild-type protein. SnRK1-overexpressing barley plants display enhanced disease resistance to Bgh. Transient expression of SnRK1 reduces fungal haustorium formation in barley cells, which probably requires SnRK1 nuclear localization and kinase activity. Together, these findings suggest that SnRK1 is directly involved in plant immunity through phosphorylation and destabilization of the WRKY3 repressor, revealing a new regulatory mechanism of immune derepression in plants.
Keywords: SnRK1; WRKY transcription factor; immunity; phosphorylation; powdery mildew fungus.
© 2020 The Author(s).
Figures


References
-
- Baena-Gonzalez E., Hanson J. Shaping plant development through the SnRK1-TOR metabolic regulators. Curr. Opin. Plant Biol. 2017;35:152–157. - PubMed
-
- Baena-Gonzalez E., Rolland F., Thevelein J.M., Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

