Maintenance of Species Differences in Closely Related Tetraploid Parasitic Euphrasia (Orobanchaceae) on an Isolated Island
- PMID: 33367265
- PMCID: PMC7748025
- DOI: 10.1016/j.xplc.2020.100105
Maintenance of Species Differences in Closely Related Tetraploid Parasitic Euphrasia (Orobanchaceae) on an Isolated Island
Abstract
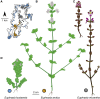
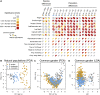
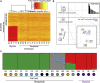
Polyploidy is pervasive in angiosperm evolution and plays important roles in adaptation and speciation. However, polyploid groups are understudied due to complex sequence homology, challenging genome assembly, and taxonomic complexity. Here, we study adaptive divergence in taxonomically complex eyebrights (Euphrasia), where recent divergence, phenotypic plasticity, and hybridization blur species boundaries. We focus on three closely related tetraploid species with contrasting ecological preferences that are sympatric on Fair Isle, a small isolated island in the British Isles. Using a common garden experiment, we show a genetic component to the morphological differences present between these species. Using whole-genome sequencing and a novel k-mer approach we call "Tetmer", we demonstrate that the species are of allopolyploid origin, with a sub-genome divergence of approximately 5%. Using ∼2 million SNPs, we show sub-genome homology across species, with a very low sequence divergence characteristic of recent speciation. This genetic variation is broadly structured by species, with clear divergence of Fair Isle heathland Euphrasia micrantha, while grassland Euphrasia arctica and coastal Euphrasia foulaensis are more closely related. Overall, we show that tetraploid Euphrasia is a system of allopolyploids of postglacial species divergence, where adaptation to novel environments may be conferred by old variants rearranged into new genetic lineages.
Keywords: allopolyploidy; divergence with gene flow; incipient speciation; k-mer spectrum; taxonomic complexity; tetraploid.
© 2020 The Authors.
Figures




Comment in
-
Plant Evolutionary Adaptation.Plant Commun. 2020 Oct 31;1(6):100118. doi: 10.1016/j.xplc.2020.100118. eCollection 2020 Nov 9. Plant Commun. 2020. PMID: 33367271 Free PMC article. No abstract available.
-
A New Year's spotlight on two years of publication.Plant Commun. 2021 Dec 29;3(1):100274. doi: 10.1016/j.xplc.2021.100274. eCollection 2022 Jan 10. Plant Commun. 2021. PMID: 35059635 Free PMC article. No abstract available.
References
-
- Abbott R.J., Brochmann C. History and evolution of the arctic flora: in the footsteps of Eric Hultén. Mol. Ecol. 2003;12:299–313. - PubMed
-
- Ainouche M.L., Fortune P.M., Salmon A., Parisod C., Grandbastien M.-A., Fukunaga K., Ricou M., Misset M.-T. Hybridization, polyploidy and invasion: lessons from Spartina (Poaceae) Biol. Invasions. 2009;11:1159–1173.
-
- Ainouche M., Chelaifa H., Ferreira J., Bellot S., Ainouche A., Salmon A. Polyploid evolution in Spartina: dealing with highly redundant hybrid genomes. In: Soltis P.S., Soltis D.E., editors. Polyploidy and Genome Evolution. Springer Berlin Heidelberg; Berlin, Heidelberg: 2012. pp. 225–243.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

