Urinary miRNA Biomarkers of Drug-Induced Kidney Injury and Their Site Specificity Within the Nephron
- PMID: 33367795
- PMCID: PMC7916737
- DOI: 10.1093/toxsci/kfaa181
Urinary miRNA Biomarkers of Drug-Induced Kidney Injury and Their Site Specificity Within the Nephron
Abstract
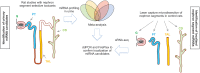
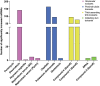
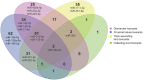
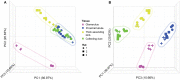
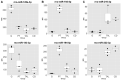
Drug-induced kidney injury (DIKI) is a major concern in both drug development and clinical practice. There is an unmet need for biomarkers of glomerular damage and more distal renal injury in the loop of Henle and the collecting duct (CD). A cross-laboratory program to identify and characterize urinary microRNA (miRNA) patterns reflecting tissue- or pathology-specific DIKI was conducted. The overall goal was to propose miRNA biomarker candidates for DIKI that could supplement information provided by protein kidney biomarkers in urine. Rats were treated with nephrotoxicants causing injury to distinct nephron segments: the glomerulus, proximal tubule, thick ascending limb (TAL) of the loop of Henle and CD. Meta-analysis identified miR-192-5p as a potential proximal tubule-specific urinary miRNA candidate. This result was supported by data obtained in laser capture microdissection nephron segments showing that miR-192-5p expression was enriched in the proximal tubule. Discriminative miRNAs including miR-221-3p and -222-3p were increased in urine from rats treated with TAL versus proximal tubule toxicants in accordance with their expression localization in the kidney. Urinary miR-210-3p increased up to 40-fold upon treatment with TAL toxicants and was also enriched in laser capture microdissection samples containing TAL and/or CD versus proximal tubule. miR-23a-3p was enriched in the glomerulus and was increased in urine from rats treated with doxorubicin, a glomerular toxicant, but not with toxicants affecting other nephron segments. Taken together these results suggest that urinary miRNA panels sourced from specific nephron regions may be useful to discriminate the pathology of toxicant-induced lesions in the kidney, thereby contributing to DIKI biomarker development needs for industry, clinical, and regulatory use.
Keywords: biomarkers; kidney injury; microRNAs; safety assessment.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society of Toxicology.
Figures



References
-
- Bauchet A. L., Masson R., Guffroy M., Slaoui M. (2011). Immunohistochemical identification of kidney nephron segments in the dog, rat, mouse, and cynomolgus monkey. Toxicol. Pathol. 39, 1115–1128. - PubMed
-
- Betton G. R., Ennulat D., Hoffman D., Gautier J. C., Harpur E., Pettit S. (2012). Biomarkers of collecting duct injury in Han-Wistar and Sprague-Dawley rats treated with N-phenylanthranilic acid. Toxicol. Pathol. 40, 682–694. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

