Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin
- PMID: 33368089
- PMCID: PMC7869224
- DOI: 10.1055/s-0040-1721319
Heparin Inhibits Cellular Invasion by SARS-CoV-2: Structural Dependence of the Interaction of the Spike S1 Receptor-Binding Domain with Heparin
Abstract
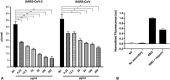
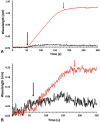
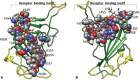
The dependence of development and homeostasis in animals on the interaction of hundreds of extracellular regulatory proteins with the peri- and extracellular glycosaminoglycan heparan sulfate (HS) is exploited by many microbial pathogens as a means of adherence and invasion. Heparin, a widely used anticoagulant drug, is structurally similar to HS and is a common experimental proxy. Exogenous heparin prevents infection by a range of viruses, including S-associated coronavirus isolate HSR1. Here, we show that heparin inhibits severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) invasion of Vero cells by up to 80% at doses achievable through prophylaxis and, particularly relevant, within the range deliverable by nebulisation. Surface plasmon resonance and circular dichroism spectroscopy demonstrate that heparin and enoxaparin, a low-molecular-weight heparin which is a clinical anticoagulant, bind and induce a conformational change in the spike (S1) protein receptor-binding domain (S1 RBD) of SARS-CoV-2. A library of heparin derivatives and size-defined fragments were used to probe the structural basis of this interaction. Binding to the RBD is more strongly dependent on the presence of 2-O or 6-O sulfate groups than on N-sulfation and a hexasaccharide is the minimum size required for secondary structural changes to be induced in the RBD. It is likely that inhibition of viral infection arises from an overlap between the binding sites of heparin/HS on S1 RBD and that of the angiotensin-converting enzyme 2. The results suggest a route for the rapid development of a first-line therapeutic by repurposing heparin and its derivatives as antiviral agents against SARS-CoV-2 and other members of the Coronaviridae.
Thieme. All rights reserved.
Conflict of interest statement
None declared.
Figures





References
-
- Ori A, Wilkinson M C, Fernig D G. The heparanome and regulation of cell function: structures, functions and challenges. Front Biosci. 2008;13:4309–4338. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

