Neuron type-specific increase in lamin B1 contributes to nuclear dysfunction in Huntington's disease
- PMID: 33369245
- PMCID: PMC7863407
- DOI: 10.15252/emmm.202012105
Neuron type-specific increase in lamin B1 contributes to nuclear dysfunction in Huntington's disease
Abstract
Lamins are crucial proteins for nuclear functionality. Here, we provide new evidence showing that increased lamin B1 levels contribute to the pathophysiology of Huntington's disease (HD), a CAG repeat-associated neurodegenerative disorder. Through fluorescence-activated nuclear suspension imaging, we show that nucleus from striatal medium-sized spiny and CA1 hippocampal neurons display increased lamin B1 levels, in correlation with altered nuclear morphology and nucleocytoplasmic transport disruption. Moreover, ChIP-sequencing analysis shows an alteration of lamin-associated chromatin domains in hippocampal nuclei, accompanied by changes in chromatin accessibility and transcriptional dysregulation. Supporting lamin B1 alterations as a causal role in mutant huntingtin-mediated neurodegeneration, pharmacological normalization of lamin B1 levels in the hippocampus of the R6/1 mouse model of HD by betulinic acid administration restored nuclear homeostasis and prevented motor and cognitive dysfunction. Collectively, our work points increased lamin B1 levels as a new pathogenic mechanism in HD and provides a novel target for its intervention.
Keywords: LAD; R6/1 mouse; chromatin accessibility; nuclear morphology; nuclear permeability.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

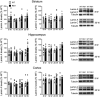
Quantification and representative immunoblots of lamin B1 in the striatum (8w, 12w, and 20w: N = 6 for both genotypes; 30w: N = 4 and 5 for WT and R6/1 mice, respectively), hippocampus (8w: N = 8 and 6 for WT and R6/1 mice, respectively; 12w: N = 7 and 6 for WT and R6/1 mice, respectively; 20w: N = 5 and 6 for WT and R6/1 mice, respectively; 30w: N = 8 and 5 for WT and R6/1 mice, respectively), and cortex (8w: N = 5 and 4 for WT and R6/1 mice, respectively; 12w and 20w: N = 6 for both genotypes; 30w: N = 6 and 5 for WT and R6/1 mice, respectively).
Quantification and representative immunoblots of lamin B2 in the striatum (8w, 12w, and 20w: N = 6 for both genotypes, 30w: N = 5 for both genotypes), hippocampus (8w: N = 8 and 6 for WT and R6/1 mice, respectively; 12w: N = 6 and 7 for WT and R6/1 mice, respectively; 20w: N = 5 for both genotypes; 30w: N = 8 and 5 for WT and R6/1 mice, respectively), and cortex (8w and 12w: N = 6 for both genotypes; 20w: N = 5 for both genotypes; 30w: N = 8 and 5 for WT and R6/1 mice, respectively).
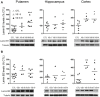
Quantification and representative immunoblots of lamin B1. Putamen: CTL N = 14; VS I‐II N = 4; VS III‐IV N = 7; hippocampus: CTL N = 9; VS I‐II N = 4; VS III‐IV N = 8; cortex: CTL N = 8; VS I‐II N = 4; VS III‐IV N = 4.
Quantification and representative immunoblots of lamin B2. Putamen: CTL N = 15; VS I‐II N = 4; VS III‐IV N = 7; hippocampus: CTL N = 6; VS I‐II N = 4; VS III‐IV N = 4; cortex: CTL N = 8; VS I‐II N = 3; VS III‐IV N = 4.
STHdhQ7/Q7 cells were transfected with a PKCδ or scrambled siRNA and PKCδ levels examined by Western blot 24 h after transfection. Graph shows the quantification (scramble N = 5 and siRNA PKCδ N = 5). A representative immunoblot is shown.
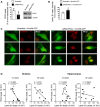
STHdhQ7/Q7 cells were transfected with a scramble or PKCδ siRNA plus N‐mHtt‐CFP and lamin B1 intensity analyzed by immunocytochemistry 24 h after transfection. Graph shows the quantification (N = 3; an average of 20 nuclei were examined for each cell culture and condition).
Representative images showing lamin B1 intensity in STHdhQ7/Q7 cells co‐transfected with N‐mHtt‐CFP (in green) and a PKCδ siRNA or a scramble siRNA. Scale bar 10 µm.
Correlation between PKCδ and lamin B1 protein levels in the striatum and hippocampus of 12‐ and 30‐week‐old WT (dark circles) and R6/1 (dark squares) mice determined by simple linear regression. Analysis was performed by using the R‐squared. Slope significantly non‐zero as indicated by *P < 0.05; ***P < 0.01. Striatum: 12 weeks: N = 6 for both genotypes; 30 weeks: N = 4 and N = 5 for WT and R6/1 mice, respectively. Hippocampus: 12 weeks: N = 7 for both genotypes; 30 weeks: N = 6 and N = 4 for WT and R6/1 mice, respectively.
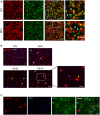
Mouse brain tissue was processed for immunohistochemistry by combining anti‐lamin B1 (red) and anti‐NeuN (green) antibodies. Representative images (maximal Z‐projections) showing the distribution of lamin B1 in the striatum of 30‐week‐old wild‐type (WT) and R6/1 mice. Yellow arrowheads show co‐localization between lamin B1 and NeuN, and white arrowheads show NeuN negative lamin B1‐positive nuclei. Scale bar 50 and 25 µm for low and high magnification, respectively.
Lamin B1 distribution in human putamen was analyzed by immunohistochemistry. Antibody against lamin B1 (red) was combined with DAPI Fluoromount‐G (blue) to label nuclei. Representative images show the distribution of lamin B1 in the putamen of non‐affected individuals (CTL) and HD patients at different stages of the disease (VS II‐IV: Vonsattel grades). Yellow and white arrowheads indicate MSNs and glial cells, respectively. Scale bar 50 and 20 µm for low and high magnification, respectively.
The distribution of lamin B1 in the putamen of HD patients was analyzed by immunohistochemistry. Anti‐lamin B1 antibody (red) was combined with anti‐GFAP antibody (green), and nuclei were labeled with DAPI Fluoromount‐G (blue). Representative images show the distribution of lamin B1 at Vonsattel grade III. Yellow arrowheads indicate MSNs, and white arrowheads indicate GFAP‐positive cells. Scale bar 10 µm.

Representative images showing the different steps of the analysis, from Z‐stack projection to nuclear segmentation, by using ImageJ.
Bar graphs showing different morphological parameters measured in wild‐type (WT, N = 3) and R6/1 (N = 3) mouse striatal nuclei. An average of 400 nuclei were examined for each sample. Bars represent the mean ± SEM. ** P < 0.01 and *P < 0.05 as compared with WT mice (two‐tailed unpaired Student’s t‐test). Exact P values are reported in Appendix Table S4.

Representative images showing the distribution of lamin B1. On the left, images showing maximal Z‐projection. Scale bar 150 µm. Small images correspond to CA1 and DG magnified images showing independent DAPI, GFAP, and lamin B1 channels, and merge, of representative confocal Z‐stack images. Scale bar 50 µm.
Left, representative images showing the distribution of lamin B1 in the nuclei of CA‐1 hippocampal neurons from WT and R6/1 mice along different Z‐axis planes (1‐6). On the right, a 3D reconstruction of the Z‐stack confocal images generated using ImageJ. Scale bar 20 µm.

Graphs show the quantification of different parameters in mice striatal MSN nuclei (Ctip2+/NeuN+). N = 4 for both genotypes. An average of 5,000 nuclei were analyzed for each sample. Representative images are shown. Scale bar 7 µm.
Graphs show the quantification of different parameters in mice striatal interneurons (Ctip2‐/NeuN+). N = 4 for both genotypes. An average of 5,000 nuclei were analyzed for each sample. Representative images are shown. Scale bar 7 µm.
Graphs show the quantification of lamin B1 intensity and circularity in 30‐week‐old R6/1 mouse striatal neuronal nuclei with (mHtt +) or without (mHtt −) nuclear inclusions. N = 6. EM48 antibody was used to label mHtt inclusions. Representative images are shown. Scale bar 7 µm.
Graphs showing lamin B1 intensity and circularity in MSN nuclei from the putamen of HD patients at different stages of the disease (VS: Vonsattel grade) and corresponding controls (CTL: non‐affected individuals). N = 6 for CTL, N = 3 for VS I‐II, and N = 4 for VS III‐IV. An average of 50 nuclei were examined for each sample. Representative images (maximal Z‐projections) are shown. Scale bar 7 µm.
Graphs show the quantification of different parameters in hippocampal CA1 (Ctip2+/Prox1−) and DG (Ctip2+/Prox1+) neuronal nuclei. N = 4 for each genotype. Representative images are shown. Scale bar 7 µm.
Graphs show the quantification of lamin B1 intensity and circularity. N = 3. An average of 12 nuclei were examined in each culture. Data are expressed as a percentage of controls. Bars represent the mean ± SEM. *P < 0.05 and **P < 0.01 as compared to mApple‐C1 control neurons (two‐tailed unpaired Student’s t‐test). Exact P values are reported in Appendix Table S3.
Representative images showing primary striatal neurons transfected with mApple‐C1 or with mApple‐Lamin B1, both in red. Neuronal nuclei were stained with DAPI Fluoromount‐G (blue). Lamin B1 is shown in white. Scale bar 10 µm.

- A
Scheme showing the experimental approach followed to measure nuclear permeability by FRAP.
- B–D
Nuclear permeability in 30‐week‐old wild‐type (WT) and R6/1 mice striatal MSNs, CA1, and DG neuronal nuclei, respectively. (B and C) N = 4 for each condition; (D) N = 5 for each condition. An average of 25 nuclei were analyzed for each sample.

UCSC genome browser capture of lamin B1 ChIP‐seq signal (log(LB1/Input)) and LADs discovered in wild‐type (WT LAD) and R6/1 (R6/1 LADs) mice in combination with NPC lamin B1 DamID (top). Black arrows highlight differences in identified LADs between WT and R6/1 mice ChIP‐seq data. Box plots of LAD size (log10(LAD size + 1)) and of LAD score (log10(LAD score + 1)) obtained by EDD (bottom) from WT (N = 3) and R6/1 (N = 3) lamin B1 ChIP‐seq data. Hipp, hippocampus. The bottom and top of the boxes are the first and third quartiles, and the line within represents the median. The whiskers denote the interval within 1.5 times the interquartile range (IQR) from the median.
Venn diagram of overlapping LADs between wild‐type (WT, N = 3) and R6/1 (N = 3) mice hippocampus (top). Metaprofile of lamin B1 and input datasets mean read density within common LADs for WT and R6/1 mice (bottom).
UCSC genome browser capture of lamin B1 ChIP‐seq signal (log(LB1/Input)) LADs discovered in wild‐type (WT LADs) and R6/1 (R6/1 LADs) mice; and common (green), wild‐type (WT)‐specific (WT spec. LADs, red), and R6/1‐specific (R6/1 spec. LADs, blue) LADs identified by EDD (top, WT (N = 3), R6/1 (N = 3)). Black arrows highlight WT‐specific LADs not identified in R6/1 mice. Box plot of average size (log10(LAD size + 1)) for common, WT‐specific, and R6/1‐specific LADs (right). The bottom and top of the boxes are the first and third quartiles, and the line within represents the median. The whiskers denote the interval within 1.5 times the interquartile range (IQR) from the median.
Lamin B1 enrichment in common, wild‐type (WT, N = 3)‐specific (spec), and R6/1 (N = 3)‐specific LADs (log10(LB1/Input reads + 1) in hippocampus (Hipp; left). Bar graphs of significant (Benjamini’s adjusted P‐value < 0.05) Biological Processes terms from DAVID for genes within common and WT‐specific LADs (right). Gene‐term enrichment was estimated by DAVID using a modified Fisher’s exact test and Benjamini’s multiple correction test. Bars represent the –log10 (Benjamini’s adjusted P‐value). Exact P values are reported in Appendix Table S3.
Representative immunoblot showing lamin B1 levels in the nucleoplasm (Nucleop) and chromatin (Chrom) in the hippocampus of 30‐week‐old wild‐type (WT, N = 7) and R6/1 (N = 7) mice. TBP, TATA‐binding protein.

Scheme showing major steps of ATAC‐seq technique.
UCSC genome browser capture of wild‐type (WT, N = 3) and R6/1 (N = 3) mice hippocampus ATAC‐seq data, hippocampal H3K9ac, and unchanged, closed in R6/1, and open in R6/1 accessible detected regions (left) in Tuba1α, Gabra2α, and Dlx2 gene locus. Arrows in blue (closed in R6/1) and red (open in R6/1) indicate differential accessible regions. Bar graphs of significant (Benjamini’s adjusted P‐value < 0.05) Biological Processes terms from DAVID for genes associated with decreased (right top) or increased (right bottom) chromatin accessibility regions. Gene‐term enrichment estimated by DAVID using a modified Fisher’s exact test and Benjamini’s multiple correction test. Bars represent the –log10 (Benjamini’s adjusted P‐value). Exact P values are reported in Appendix Table S3.
Box plots showing average gene expression (log10(FPKMs + 1) for genes up‐ or down‐regulated (adjusted P‐value < 0.001, |FC|>2) in R6/1 (N = 9) versus WT (N = 9) mice (left). The bottom and top of the boxes are the first and third quartiles, and the line within represents the median. The whiskers denote the interval within 1.5 times the interquartile range (IQR) from the median. Heat map showing expression profile (log10 (FPKMs + 1)) of genes up‐ or down‐regulated (adjusted P‐value < 0.001, |FC|>2, N = 9) in R6/1 mice (mid). Genes are ranked by the degree of expression. Numbers in color scale show the correspondence between gene expression values and colors. Box plots showing average TSS chromatin accessibility (log10(ATAC reads + 1), N = 3) for genes up‐ or down‐regulated (adjusted P‐value < 0.001, |FC|>2, N = 9) in R6/1 versus WT mice hippocampus (right). Exact P values are reported in Appendix Table S3. The bottom and top of the boxes are the first and third quartiles, and the line within represents the median. The whiskers denote the interval within 1.5 times the interquartile range (IQR) from the median.
Average gene expression (closed or open regions FPKMs/ unchanged regions FPKMs) for genes associated with differential accessible regions in R6/1 (N = 9) versus WT (N = 9) mice (adjusted P‐value < 0.05, N = 3). Each point corresponds to the value from an individual sample. Data are shown as the mean ± SEM. *P < 0.05 as compared with WT mice (two‐tailed unpaired Student’s t‐test). Exact P values are reported in Appendix Table S3.

Box plots of average expression (log10(FPKMs + 1)) for genes sublists 1‐5 (lowest to highest expression) for wild‐type (WT, N = 3) mice (left). Pie charts of gene distribution among generated sublists (1‐5) for all genes (middle) and genes in LADs (right) in WT mice. The bottom and top of the boxes are the first and third quartiles, and the line within represents the median. The whiskers denote the interval within 1.5 times the interquartile range (IQR) from the median.
Average gene expression (FPKMs) of genes found in common and wild‐type (WT, N = 3)‐specific LADs for WT (N = 9) and R6/1 (N = 9) mice. Each point corresponds to the value from an individual sample. Data are shown as the mean ± SEM. * P < 0.05 as compared to WT mice (two‐tailed unpaired Student’s t‐test). Exact P values are reported in Appendix Table S3.
Average chromatin accessibility (normalized read counts) of genes found in common and wild‐type (WT, N = 3)‐specific LADs for WT (N = 3) and R6/1 (N = 3) mice. Each point corresponds to the value from an individual sample. Data are shown as the mean ± SEM. The Wilcoxon–Mann–Whitney test was used for statistical analysis. Exact P values are reported in Appendix Table S3.
Venn diagram showing the total number of genes found in common and wild‐type (WT)‐specific LAD regions (left). Bar graph showing the number of up‐ and down‐regulated genes found in common and WT‐specific LAD regions filtering only according to adjusted P‐value (adjusted P‐value < 0.001) or additionally with fold change (adjusted P‐value < 0.001, |FC| > 2) obtained from deseq2 differential expression analysis (see methods).
UCSC genome browser capture of wild‐type (WT, N = 3) and R6/1 (N = 3) mice hippocampus ATAC‐seq data, lamin B1 ChIP‐seq (log(Lb1 ChIP/Input)) for WT (N = 3) and R6/1 (N = 3) mice hippocampus, hippocampal H3K9ac, CTCF, and H3K9me3 for WT mice hippocampus in Fos locus (left). Enhancer regions (E1‐E5) are indicated with arrows. Box plot of lamin B1 enrichment (log10(Lb1 ChIP/Input)) for regions with unchanged, decreased (closed in R6/1), and increased (opened in R6/1) chromatin accessibility in R6/1 (N = 3) mice hippocampus. * P < 0.05 as compared to WT (N = 3) mice (the Wilcoxon–Mann–Whitney test). Exact P values are reported in Appendix Table S3. The bottom and top of the boxes are the first and third quartiles, and the line within represents the median. The whiskers denote the interval within 1.5 times the interquartile range (IQR) from the median.

- A
Timeline of the behavioral, biochemical, and histopathological analyses performed in wild‐type (WT) and R6/1 mice to assess the effect of betulinic acid (BA) administration. w, weeks; AR, accelerating rotarod.
- B, C
Graphs show the percentage of time exploring each object with respect to the total exploration time in the (B) NOLT and (C) NORT, 5 weeks after treatment (Veh, vehicle; BA, betulinic acid; WT, wild‐type). ****P < 0.0001 compared with the corresponding old location/object. WT veh N = 13; R6/1 veh N = 10; R6/1 + BA N = 10.
- D
Accelerating rotarod was assessed after 7 weeks of treatment. **P < 0.01 and ***P < 0.001 compared with vehicle‐treated wild‐type (WT) mice; # P < 0.05 compared with vehicle‐treated R6/1 mice. WT veh N = 13; R6/1 veh N = 9; R6/1 + BA N = 9.
- E
Lamin B1 levels were analyzed by Western blot in the striatum (WT veh N = 10; R6/1 veh N = 6; R6/1 + BA N = 7), hippocampus (WT veh N = 12; R6/1 veh N = 8; R6/1 + BA N = 7), and cortex (WT veh N = 11; R6/1 veh N = 6; R6/1 + BA N = 7) after 12 weeks of treatment. *P < 0.05, **P < 0.05 compared with vehicle‐treated wild‐type (WT) mice and # P < 0.05 compared with vehicle‐treated R6/1 mice. Representative immunoblots of lamin B1 and α‐tubulin (as loading control) for each treatment group are shown.
- F
Lamin B1 intensity and nuclear morphology were analyzed by FANSI in hippocampal CA1 neuronal nuclei from wild‐type (WT) and R6/1 mice after 12 weeks of treatment. *P < 0.05 compared with vehicle‐treated WT mice. *P < 0.05 as compared to vehicle‐treated WT mice. Lamin B1 intensity: WT veh N = 6; R6/1 veh N = 5; R6/1 + BA N = 4; lamin B1 circularity: WT veh N = 6; R6/1 veh N = 5; R6/1 + BA N = 5.
- G
Nuclear permeability was analyzed by FRAP in hippocampal CA1 neuronal nuclei after 12 weeks of treatment. *P < 0.05 compared with vehicle‐treated wild‐type (WT) mice. WT veh N = 4; R6/1 veh N = 5; R6/1 + BA N = 6.
References
-
- Alvarez‐Periel E, Puigdellívol M, Brito V, Plattner F, Bibb JA, Alberch J, Ginés S (2018) Cdk5 contributes to huntington’s disease learning and memory deficits via modulation of brain region‐specific substrates. Mol Neurobiol 55: 6250–6268 - PubMed
-
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, MacKlis JD (2005) Neuronal subtype‐specific genes that control corticospinal motor neuron development in vivo . Neuron 45: 207–221 - PubMed
-
- Bagri A, Gurney T, He X, Zou YR, Littman DR, Tessier‐Lavigne M, Pleasure SJ (2002) The chemokine SDF1 regulates migration of dentate granule cells. Development 129: 4249–4260 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

