Community Approaches for Integrating Environmental Exposures into Human Models of Disease
- PMID: 33369481
- PMCID: PMC7769179
- DOI: 10.1289/EHP7215
Community Approaches for Integrating Environmental Exposures into Human Models of Disease
Abstract
Background: A critical challenge in genomic medicine is identifying the genetic and environmental risk factors for disease. Currently, the available data links a majority of known coding human genes to phenotypes, but the environmental component of human disease is extremely underrepresented in these linked data sets. Without environmental exposure information, our ability to realize precision health is limited, even with the promise of modern genomics. Achieving integration of gene, phenotype, and environment will require extensive translation of data into a standard, computable form and the extension of the existing gene/phenotype data model. The data standards and models needed to achieve this integration do not currently exist.
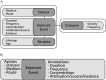
Objectives: Our objective is to foster development of community-driven data-reporting standards and a computational model that will facilitate the inclusion of exposure data in computational analysis of human disease. To this end, we present a preliminary semantic data model and use cases and competency questions for further community-driven model development and refinement.
Discussion: There is a real desire by the exposure science, epidemiology, and toxicology communities to use informatics approaches to improve their research workflow, gain new insights, and increase data reuse. Critical to success is the development of a community-driven data model for describing environmental exposures and linking them to existing models of human disease. https://doi.org/10.1289/EHP7215.
Figures


References
-
- Aaseth J, Wallace DR, Vejrup K, Alexander J. 2020. Methylmercury and developmental neurotoxicity: a global concern. Curr Opin Toxicol 19:80–87, 10.1016/j.cotox.2020.01.005. - DOI
-
- Boyles RR, Thessen AE, Waldrop A, Haendel MA. 2019. Ontology-based data integration for advancing toxicological knowledge. Curr Opin Toxicol 16:67–74, 10.1016/j.cotox.2019.05.005. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
