Multi-tissue transcriptome-wide association studies
- PMID: 33369784
- PMCID: PMC8048510
- DOI: 10.1002/gepi.22374
Multi-tissue transcriptome-wide association studies
Abstract
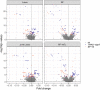
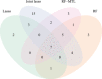
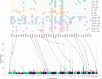
A transcriptome-wide association study (TWAS) attempts to identify disease associated genes by imputing gene expression into a genome-wide association study (GWAS) using an expression quantitative trait loci (eQTL) data set and then testing for associations with a trait of interest. Regulatory processes may be shared across related tissues and one natural extension of TWAS is harnessing cross-tissue correlation in gene expression to improve prediction accuracy. Here, we studied multi-tissue extensions of lasso regression and random forests (RF), joint lasso and RF-MTL (multi-task learning RF), respectively. We found that, on our chosen eQTL data set, multi-tissue methods were generally more accurate than their single-tissue counterparts, with RF-MTL performing the best. Simulations showed that these benefits generally translated into more associated genes identified, although highlighted that joint lasso had a tendency to erroneously identify genes in one tissue if there existed an eQTL signal for that gene in another. Applying the four methods to a type 1 diabetes GWAS, we found that multi-tissue methods found more unique associated genes for most of the tissues considered. We conclude that multi-tissue methods are competitive and, for some cell types, superior to single-tissue approaches and hold much promise for TWAS studies.
Keywords: complex traits; gene expression; multi-task learning; transcriptome-wide association studies.
© 2020 The Authors. Genetic Epidemiology published by Wiley Periodicals LLC.
Figures


References
-
- Balasubramanian, K. , Yu, K. , & Zhang, T. (2013). High‐dimensional joint sparsity random effects model for multi‐task learning. ArXiv:1309.6814 [Cs, Stat]. http://arxiv.org/abs/1309.6814
-
- Barrett, J. C. , Clayton, D. G. , Concannon, P. , Akolkar, B. , Cooper, J. D. , & Erlich, H. A. , Type 1 Diabetes Genetics Consortium . (2009). Genome‐wide association study and meta‐analysis find that over 40 loci affect risk of type 1 diabetes. Nature Genetics, 41(6), 703–707. 10.1038/ng.381 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

