Genetic variation in the Middle East-an opportunity to advance the human genetics field
- PMID: 33371902
- PMCID: PMC7768658
- DOI: 10.1186/s13073-020-00821-7
Genetic variation in the Middle East-an opportunity to advance the human genetics field
Abstract
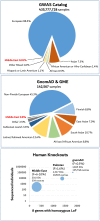
We highlight the current lack of representation of the Middle East from large genomic studies and emphasize the expected high impact of cataloging its variation. We discuss the limiting factors and possible solutions to generating and accessing research and clinical sequencing data from this part of the world.
Conflict of interest statement
Rehm declares a pending NIH grant from the National Human Genome Research Institute to support gnomAD. The remaining author declares that he has no competing interests.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources

