Deep learning for in vivo near-infrared imaging
- PMID: 33372162
- PMCID: PMC7817119
- DOI: 10.1073/pnas.2021446118
Deep learning for in vivo near-infrared imaging
Abstract
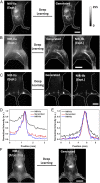
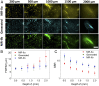
Detecting fluorescence in the second near-infrared window (NIR-II) up to ∼1,700 nm has emerged as a novel in vivo imaging modality with high spatial and temporal resolution through millimeter tissue depths. Imaging in the NIR-IIb window (1,500-1,700 nm) is the most effective one-photon approach to suppressing light scattering and maximizing imaging penetration depth, but relies on nanoparticle probes such as PbS/CdS containing toxic elements. On the other hand, imaging the NIR-I (700-1,000 nm) or NIR-IIa window (1,000-1,300 nm) can be done using biocompatible small-molecule fluorescent probes including US Food and Drug Administration-approved dyes such as indocyanine green (ICG), but has a caveat of suboptimal imaging quality due to light scattering. It is highly desired to achieve the performance of NIR-IIb imaging using molecular probes approved for human use. Here, we trained artificial neural networks to transform a fluorescence image in the shorter-wavelength NIR window of 900-1,300 nm (NIR-I/IIa) to an image resembling an NIR-IIb image. With deep-learning translation, in vivo lymph node imaging with ICG achieved an unprecedented signal-to-background ratio of >100. Using preclinical fluorophores such as IRDye-800, translation of ∼900-nm NIR molecular imaging of PD-L1 or EGFR greatly enhanced tumor-to-normal tissue ratio up to ∼20 from ∼5 and improved tumor margin localization. Further, deep learning greatly improved in vivo noninvasive NIR-II light-sheet microscopy (LSM) in resolution and signal/background. NIR imaging equipped with deep learning could facilitate basic biomedical research and empower clinical diagnostics and imaging-guided surgery in the clinic.
Keywords: deep learning; near-infrared imaging; second near-infrared window.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Hong G., et al. , In vivo fluorescence imaging with Ag2S quantum dots in the second near-infrared region. Angew. Chem. Int. Ed. Engl. 51, 9818–9821 (2012). - PubMed
-
- Hong G., et al. , Ultrafast fluorescence imaging in vivo with conjugated polymer fluorophores in the second near-infrared window. Nat. Commun. 5, 4206 (2014). - PubMed
-
- Antaris A. L., et al. , A small-molecule dye for NIR-II imaging. Nat. Mater. 15, 235–242 (2016). - PubMed
-
- Cosco E. D., et al. , Flavylium polymethine fluorophores for near- and shortwave infrared imaging. Angew. Chem. Int. Ed. Engl. 56, 13126–13129 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

