Single-cell RNA sequencing reveals regulation of fetal ovary development in the monkey (Macaca fascicularis)
- PMID: 33372178
- PMCID: PMC7769980
- DOI: 10.1038/s41421-020-00219-0
Single-cell RNA sequencing reveals regulation of fetal ovary development in the monkey (Macaca fascicularis)
Abstract
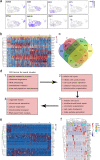
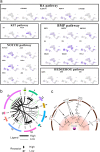
Germ cells are vital for reproduction and heredity. However, the mechanisms underlying female germ cell development in primates, especially in late embryonic stages, remain elusive. Here, we performed single-cell RNA sequencing of 12,471 cells from whole fetal ovaries, and explored the communications between germ cells and niche cells. We depicted the two waves of oogenesis at single-cell resolution and demonstrated that progenitor theca cells exhibit similar characteristics to Leydig cells in fetal monkey ovaries. Notably, we found that ZGLP1 displays differentially expressed patterns between mouse and monkey, which is not overlapped with NANOG in monkey germ cells, suggesting its role in meiosis entry but not in activating oogenic program in primates. Furthermore, the majority of germ cell clusters that sharply express PRDM9 and SPO11 might undergo apoptosis after cyst breakdown, leading to germ cell attrition. Overall, our work provides new insights into the molecular and cellular basis of primate fetal ovary development at single-cell resolution.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Stévant, I. et al. Single-cell transcriptomics of the mouse gonadal soma reveals the establishment of sexual dimorphism in distinct cell lineages. https://www.biorxiv.org/content/10.1101/410407v1.full (2018). - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

