Identification of a cytosine methyltransferase that improves transformation efficiency in Methylomonas sp. DH-1
- PMID: 33372613
- PMCID: PMC7720504
- DOI: 10.1186/s13068-020-01846-1
Identification of a cytosine methyltransferase that improves transformation efficiency in Methylomonas sp. DH-1
Abstract
Background: Industrial biofuels and other value-added products can be produced from metabolically engineered microorganisms. Methylomonas sp. DH-1 is a candidate platform for bioconversion that uses methane as a carbon source. Although several genetic engineering techniques have been developed to work with Methylomonas sp. DH-1, the genetic manipulation of plasmids remains difficult because of the restriction-modification (RM) system present in the bacteria. Therefore, the RM system in Methylomonas sp. DH-1 must be identified to improve the genetic engineering prospects of this microorganism.
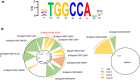
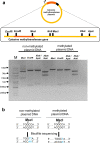
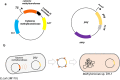
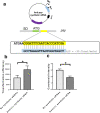
Results: We identified a DNA methylation site, TGGCCA, and its corresponding cytosine methyltransferase for the first time in Methylomonas sp. DH-1 through whole-genome bisulfite sequencing. The methyltransferase was confirmed to methylate the fourth nucleotide of TGGCCA. In general, methylated plasmids exhibited better transformation efficiency under the protection of the RM system than non-methylated plasmids did. As expected, when we transformed Methylomonas sp. DH-1 with plasmid DNA harboring the psy gene, the metabolic flux towards carotenoid increased. The methyltransferase-treated plasmid exhibited an increase in transformation efficiency of 2.5 × 103 CFU/μg (124%). The introduced gene increased the production of carotenoid by 26%. In addition, the methyltransferase-treated plasmid harboring anti-psy sRNA gene exhibited an increase in transformation efficiency by 70% as well. The production of carotenoid was decreased by 40% when the psy gene was translationally repressed by anti-psy sRNA.
Conclusions: Plasmid DNA methylated by the discovered cytosine methyltransferase from Methylomonas sp. DH-1 had a higher transformation efficiency than non-treated plasmid DNA. The RM system identified in this study may facilitate the plasmid-based genetic manipulation of methanotrophs.
Keywords: Cytosine methyltransferase; DNA methylation; Methylomonas sp. DH-1; Transformation efficiency.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Construction of a tunable promoter library to optimize gene expression in Methylomonas sp. DH-1, a methanotroph, and its application to cadaverine production.Biotechnol Biofuels. 2021 Dec 4;14(1):228. doi: 10.1186/s13068-021-02077-8. Biotechnol Biofuels. 2021. PMID: 34863247 Free PMC article.
-
Advanced biotechnology using methyltransferase and its applications in bacteria: a mini review.Biotechnol Lett. 2022 Jan;44(1):33-44. doi: 10.1007/s10529-021-03208-9. Epub 2021 Nov 25. Biotechnol Lett. 2022. PMID: 34820721 Review.
-
A comparative transcriptome analysis of the novel obligate methanotroph Methylomonas sp. DH-1 reveals key differences in transcriptional responses in C1 and secondary metabolite pathways during growth on methane and methanol.BMC Genomics. 2019 Feb 12;20(1):130. doi: 10.1186/s12864-019-5487-6. BMC Genomics. 2019. PMID: 30755173 Free PMC article.
-
Metabolic engineering of the type I methanotroph Methylomonas sp. DH-1 for production of succinate from methane.Metab Eng. 2019 Jul;54:170-179. doi: 10.1016/j.ymben.2019.03.013. Epub 2019 Apr 12. Metab Eng. 2019. PMID: 30986511
-
Genetic manipulation of Staphylococci-breaking through the barrier.Front Cell Infect Microbiol. 2012 Apr 12;2:49. doi: 10.3389/fcimb.2012.00049. eCollection 2012. Front Cell Infect Microbiol. 2012. PMID: 22919640 Free PMC article. Review.
Cited by
-
Emerging methylation-based approaches in microbiome engineering.Biotechnol Biofuels Bioprod. 2024 Jul 10;17(1):96. doi: 10.1186/s13068-024-02529-x. Biotechnol Biofuels Bioprod. 2024. PMID: 38987811 Free PMC article. Review.
-
Bacterial DNA methylases as novel molecular and synthetic biology tools: recent developments.Appl Microbiol Biotechnol. 2025 Mar 6;109(1):60. doi: 10.1007/s00253-025-13442-0. Appl Microbiol Biotechnol. 2025. PMID: 40047928 Free PMC article. Review.
-
Construction of a tunable promoter library to optimize gene expression in Methylomonas sp. DH-1, a methanotroph, and its application to cadaverine production.Biotechnol Biofuels. 2021 Dec 4;14(1):228. doi: 10.1186/s13068-021-02077-8. Biotechnol Biofuels. 2021. PMID: 34863247 Free PMC article.
-
Advanced biotechnology using methyltransferase and its applications in bacteria: a mini review.Biotechnol Lett. 2022 Jan;44(1):33-44. doi: 10.1007/s10529-021-03208-9. Epub 2021 Nov 25. Biotechnol Lett. 2022. PMID: 34820721 Review.
-
Advances in Diversity, Evolutionary Dynamics and Biotechnological Potential of Restriction-Modification Systems.Microorganisms. 2025 May 14;13(5):1126. doi: 10.3390/microorganisms13051126. Microorganisms. 2025. PMID: 40431298 Free PMC article. Review.
References
-
- Xin JY, Cui JR, Niu JZ, Hua SF, Xia CG, Li SB, et al. Production of methanol from methane by methanotrophic bacteria. Biocatal Biotransform. 2004;22(3):225–229. doi: 10.1080/10242420412331283305. - DOI
-
- Brautaset T, Jakobsen OM, Degnes KF, Netzer R, Naerdal I, Krog A, et al. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for l-lysine production from methanol at 50 degrees C. Appl Microbiol Biotechnol. 2010;87(3):951–964. doi: 10.1007/s00253-010-2559-6. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

