Mitochondria-lysosome membrane contacts are defective in GDAP1-related Charcot-Marie-Tooth disease
- PMID: 33372681
- PMCID: PMC7823109
- DOI: 10.1093/hmg/ddaa243
Mitochondria-lysosome membrane contacts are defective in GDAP1-related Charcot-Marie-Tooth disease
Abstract
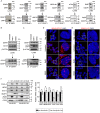
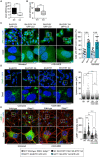
Mutations in the GDAP1 gene cause Charcot-Marie-Tooth (CMT) neuropathy. GDAP1 is an atypical glutathione S-transferase (GST) of the outer mitochondrial membrane and the mitochondrial membrane contacts with the endoplasmic reticulum (MAMs). Here, we investigate the role of this GST in the autophagic flux and the membrane contact sites (MCSs) between mitochondria and lysosomes in the cellular pathophysiology of GDAP1 deficiency. We demonstrate that GDAP1 participates in basal autophagy and that its depletion affects LC3 and PI3P biology in autophagosome biogenesis and membrane trafficking from MAMs. GDAP1 also contributes to the maturation of lysosome by interacting with PYKfyve kinase, a pH-dependent master lysosomal regulator. GDAP1 deficiency causes giant lysosomes with hydrolytic activity, a delay in the autophagic lysosome reformation, and TFEB activation. Notably, we found that GDAP1 interacts with LAMP-1, which supports that GDAP1-LAMP-1 is a new tethering pair of mitochondria and lysosome membrane contacts. We observed mitochondria-lysosome MCSs in soma and axons of cultured mouse embryonic motor neurons and human neuroblastoma cells. GDAP1 deficiency reduces the MCSs between these organelles, causes mitochondrial network abnormalities, and decreases levels of cellular glutathione (GSH). The supply of GSH-MEE suffices to rescue the lysosome membranes and the defects of the mitochondrial network, but not the interorganelle MCSs nor early autophagic events. Overall, we show that GDAP1 enables the proper function of mitochondrial MCSs in both degradative and nondegradative pathways, which could explain primary insults in GDAP1-related CMT pathophysiology, and highlights new redox-sensitive targets in axonopathies where mitochondria and lysosomes are involved.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures






References
-
- Marco A., Cuesta A., Pedrola L., Palau F. and Marin I. (2004) Evolutionary and structural analyses of GDAP1, involved in Charcot-Marie-Tooth disease, characterize a novel class of glutathione transferase-related genes. Mol. Biol. Evol., 21, 176–187. - PubMed
-
- Baxter R.V., Ben Othmane K., Rochelle J.M., Stajich J.E., Hulette C., Dew-Knight S., Hentati F., Ben Hamida M., Bel S., Stenger J.E. et al. (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat. Genet., 30, 21–22. - PubMed
-
- Cuesta A., Pedrola L., Sevilla T., Garcia-Planells J., Chumillas M.J., Mayordomo F., LeGuern E., Marin I., Vilchez J.J. and Palau F. (2002) The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat. Genet., 30, 22–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

