Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an in silico perspective
- PMID: 33372806
- PMCID: PMC7783895
- DOI: 10.1177/2040206620984076
Alkaloids and flavonoids from African phytochemicals as potential inhibitors of SARS-Cov-2 RNA-dependent RNA polymerase: an in silico perspective
Abstract
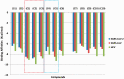
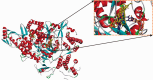
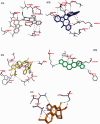
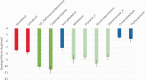
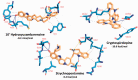
Corona Virus Disease 2019 (COVID-19) is a pandemic caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2). Exploiting the potentials of phytocompounds is an integral component of the international response to this pandemic. In this study, a virtual screening through molecular docking analysis was used to screen a total of 226 bioactive compounds from African herbs and medicinal plants for direct interactions with SARS-CoV-2 RNA-dependent RNA polymerase (RdRp). From these, 36 phytocompounds with binding affinities higher than the approved reference drugs (remdesivir and sobosivir), were further docked targeting the active sites of SARS-CoV-2, as well as SARS-CoV and HCV RdRp. A hit list of 7 compounds alongside two positive controls (remdesivir and sofosbuvir) and two negative controls (cinnamaldehyde and Thymoquinone) were further docked into the active site of 8 different conformations of SARS-CoV-2 RdRp gotten from molecular dynamics simulation (MDS) system equilibration. The top docked compounds were further subjected to predictive druglikeness and ADME/tox filtering analyses. Drugable alkaloids (10'-hydroxyusambarensine, cryptospirolepine, strychnopentamine) and flavonoids (usararotenoid A, and 12α-epi-millettosin), were reported to exhibit strong affinity binding and interactions with key amino acid residues in the catalytic site, the divalent-cation-binding site, and the NTP entry channel in the active region of the RdRp enzyme as the positive controls. These phytochemicals, in addition to other promising antivirals such as remdesivir and sofosbuvir, may be exploited towards the development of a cocktail of anti-coronavirus treatments in COVID-19. Experimental studies are recommended to validate these study.
Keywords: RNA-dependent RNA polymerase; SARS-CoV-2; alkaloids; flavonoids; phytochemicals.
Conflict of interest statement
Figures

References
-
- Porcheddu R, Serra C, Kelvin D, et al. Similarity in case fatality rates (CFR) of COVID-19/SARS-COV-2 in Italy and China. J Infect Dev Ctries 2020; 14: 125–128. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

