SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species
- PMID: 33372854
- PMCID: PMC7850364
- DOI: 10.1080/22221751.2020.1870414
SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species
Abstract
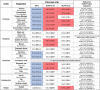
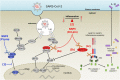
The genome of SARS-CoV-2 encodes two viral proteases (NSP3/papain-like protease and NSP5/3C-like protease) that are responsible for cleaving viral polyproteins during replication. Here, we discovered new functions of the NSP3 and NSP5 proteases of SARS-CoV-2, demonstrating that they could directly cleave proteins involved in the host innate immune response. We identified 3 proteins that were specifically and selectively cleaved by NSP3 or NSP5: IRF-3, and NLRP12 and TAB1, respectively. Direct cleavage of IRF3 by NSP3 could explain the blunted Type-I IFN response seen during SARS-CoV-2 infections while NSP5 mediated cleavage of NLRP12 and TAB1 point to a molecular mechanism for enhanced production of cytokines and inflammatory responThe genome of SARS-CoV-2 encodes two viral proteases (NSP3/papain-like protease and NSP5/3C-like protease) that are responsible for cleaving viral polyproteins during replication. Here, we discovered new functions of the NSP3 and NSP5 proteases of SARS-CoV-2, demonstrating that they could directly cleave proteins involved in the host innate immune response. We identified 3 proteins that were specifically and selectively cleaved by NSP3 or NSP5: IRF-3, and NLRP12 and TAB1, respectively. Direct cleavage of IRF3 by NSP3 could explain the blunted Type-I IFN response seen during SARS-CoV-2 infections while NSP5 mediated cleavage of NLRP12 and TAB1 point to a molecular mechanism for enhanced production of cytokines and inflammatory response observed in COVID-19 patients. We demonstrate that in the mouse NLRP12 protein, one of the recognition site is not cleaved in our in-vitro assay. We pushed this comparative alignment of IRF-3 and NLRP12 homologs and show that the lack or presence of cognate cleavage motifs in IRF-3 and NLRP12 could contribute to the presentation of disease in cats and tigers, for example. Our findings provide an explanatory framework for indepth studies into the pathophysiology of COVID-19.
Keywords: IRF3; NLRP12; NSP3 (PLpro); NSP5 (3CLpro); SARS-CoV-2; TAB1; innate immunity; protease activity.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






Similar articles
-
SARS-CoV-2 nsp5 Exhibits Stronger Catalytic Activity and Interferon Antagonism than Its SARS-CoV Ortholog.J Virol. 2022 Apr 27;96(8):e0003722. doi: 10.1128/jvi.00037-22. Epub 2022 Apr 7. J Virol. 2022. PMID: 35389264 Free PMC article.
-
SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3.Int J Biol Sci. 2021 Apr 10;17(6):1547-1554. doi: 10.7150/ijbs.59943. eCollection 2021. Int J Biol Sci. 2021. PMID: 33907518 Free PMC article.
-
SARS-CoV-2 Nsp5 Demonstrates Two Distinct Mechanisms Targeting RIG-I and MAVS To Evade the Innate Immune Response.mBio. 2021 Oct 26;12(5):e0233521. doi: 10.1128/mBio.02335-21. Epub 2021 Sep 21. mBio. 2021. PMID: 34544279 Free PMC article.
-
Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19.J Gen Virol. 2021 Mar;102(3):001558. doi: 10.1099/jgv.0.001558. Epub 2021 Jan 28. J Gen Virol. 2021. PMID: 33507143 Free PMC article. Review.
-
Spatial and temporal roles of SARS-CoV PLpro -A snapshot.FASEB J. 2021 Jan;35(1):e21197. doi: 10.1096/fj.202002271. FASEB J. 2021. PMID: 33368679 Free PMC article. Review.
Cited by
-
The subversion of toll-like receptor signaling by bacterial and viral proteases during the development of infectious diseases.Mol Aspects Med. 2022 Dec;88:101143. doi: 10.1016/j.mam.2022.101143. Epub 2022 Sep 21. Mol Aspects Med. 2022. PMID: 36152458 Free PMC article. Review.
-
Inhibition of SARS-CoV-2 3CL protease by the anti-viral chimeric protein RetroMAD1.Sci Rep. 2023 Nov 17;13(1):20178. doi: 10.1038/s41598-023-47511-z. Sci Rep. 2023. PMID: 37978223 Free PMC article.
-
The main protease of SARS-CoV-2 cleaves histone deacetylases and DCP1A, attenuating the immune defense of the interferon-stimulated genes.J Biol Chem. 2023 Mar;299(3):102990. doi: 10.1016/j.jbc.2023.102990. Epub 2023 Feb 8. J Biol Chem. 2023. PMID: 36758802 Free PMC article.
-
Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin.bioRxiv [Preprint]. 2023 Jan 19:2021.09.15.460543. doi: 10.1101/2021.09.15.460543. bioRxiv. 2023. Update in: Nat Commun. 2023 Apr 25;14(1):2366. doi: 10.1038/s41467-023-38031-5. PMID: 35547846 Free PMC article. Updated. Preprint.
-
Sensitivity to Vaccines, Therapeutic Antibodies, and Viral Entry Inhibitors and Advances To Counter the SARS-CoV-2 Omicron Variant.Clin Microbiol Rev. 2022 Sep 21;35(3):e0001422. doi: 10.1128/cmr.00014-22. Epub 2022 Jun 6. Clin Microbiol Rev. 2022. PMID: 35862736 Free PMC article. Review.
References
-
- Qiu Y, Xu K.. Functional studies of the coronavirus nonstructural proteins. STE Med. 2020;1(2):e39. 10.37175/stemedecine.v1i2.39 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
