Characterization of the Class I MHC Peptidome Resulting From DNCB Exposure of HaCaT Cells
- PMID: 33372950
- PMCID: PMC7916740
- DOI: 10.1093/toxsci/kfaa184
Characterization of the Class I MHC Peptidome Resulting From DNCB Exposure of HaCaT Cells
Abstract
Skin sensitization following the covalent modification of proteins by low molecular weight chemicals (haptenation) is mediated by cytotoxic T lymphocyte (CTL) recognition of human leukocyte antigen (HLA) molecules presented on the surface of almost all nucleated cells. There exist 3 nonmutually exclusive hypotheses for how haptens mediate CTL recognition: direct stimulation by haptenated peptides, hapten modification of HLA leading to an altered HLA-peptide repertoire, or a hapten altered proteome leading to an altered HLA-peptide repertoire. To shed light on the mechanism underpinning skin sensitization, we set out to utilize proteomic analysis of keratinocyte presented antigens following exposure to 2,4-dinitrochlorobenzene (DNCB). We show that the following DNCB exposure, cultured keratinocytes present cysteine haptenated (dinitrophenylated) peptides in multiple HLA molecules. In addition, we find that one of the DNCB modified peptides derives from the active site of cytosolic glutathione-S transferase-ω. These results support the current view that a key mechanism of skin sensitization is stimulation of CTLs by haptenated peptides. Data are available via ProteomeXchange with identifier PXD021373.
Keywords: DNCB; HLA; HaCaT; keratinocyte; peptidome.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society of Toxicology.
Figures
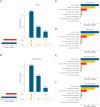

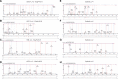
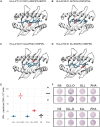
References
-
- Aleksic M., Pease C. K., Basketter D. A., Panico M., Morris H. R., Dell A. (2008). Mass spectrometric identification of covalent adducts of the skin allergen 2, 4-dinitro-1-chlorobenzene and model skin proteins. Toxicol. In Vitro 22, 1169–1176. - PubMed
-
- Aleksic M., Thain E., Roger D., Saib O., Davies M., Li J., Aptula A., Zazzeroni R. (2009). Reactivity profiling: Covalent modification of single nucleophile peptides for skin sensitization risk assessment. Toxicol. Sci. 108, 401–411. - PubMed
-
- Banerjee G., Damodaran A., Devi N., Dharmalingam K., Raman G. (2004). Role of keratinocytes in antigen presentation and polarization of human T lymphocytes. Scand. J. Immunol. 59, 385–394. - PubMed
-
- Barnstable C. J., Bodmer W. F., Brown G., Galfre G., Milstein C., Williams A. F., Ziegler A. (1978). Production of monoclonal antibodies to group a erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell 14, 9–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

