New Insights into the Ecology and Physiology of Methanomassiliicoccales from Terrestrial and Aquatic Environments
- PMID: 33374130
- PMCID: PMC7824343
- DOI: 10.3390/microorganisms9010030
New Insights into the Ecology and Physiology of Methanomassiliicoccales from Terrestrial and Aquatic Environments
Abstract
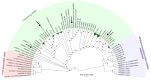
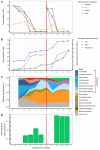
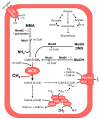
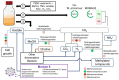
Members of the archaeal order Methanomassiliicoccales are methanogens mainly associated with animal digestive tracts. However, environmental members remain poorly characterized as no representatives not associated with a host have been cultivated so far. In this study, metabarcoding screening combined with quantitative PCR analyses on a collection of diverse non-host-associated environmental samples revealed that Methanomassiliicoccales were very scarce in most terrestrial and aquatic ecosystems. Relative abundance of Methanomassiliicoccales and substrates/products of methanogenesis were monitored during incubation of environmental slurries. A sediment slurry enriched in Methanomassiliicoccales was obtained from a freshwater sample. It allowed the reconstruction of a high-quality metagenome-assembled genome (MAG) corresponding to a new candidate species, for which we propose the name of Candidatus 'Methanomassiliicoccus armoricus MXMAG1'. Comparison of the annotated genome of MXMAG1 with the published genomes and MAGs from Methanomassiliicoccales belonging to the 2 known clades ('free-living'/non-host-associated environmental clade and 'host-associated'/digestive clade) allowed us to explore the putative physiological traits of Candidatus 'M. armoricus MXMAG1'. As expected, Ca. 'Methanomassiliicoccus armoricus MXMAG1' had the genetic potential to produce methane by reduction of methyl compounds and dihydrogen oxidation. This MAG encodes for several putative physiological and stress response adaptations, including biosynthesis of trehalose (osmotic and temperature regulations), agmatine production (pH regulation), and arsenic detoxication, by reduction and excretion of arsenite, a mechanism that was only present in the 'free-living' clade. An analysis of co-occurrence networks carried out on environmental samples and slurries also showed that Methanomassiliicoccales detected in terrestrial and aquatic ecosystems were strongly associated with acetate and dihydrogen producing bacteria commonly found in digestive habitats and which have been reported to form syntrophic relationships with methanogens.
Keywords: Methanomassiliicoccales; cultivation; environmental cluster; methyl-compounds; networks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kirschke S., Bousquet P., Ciais P., Saunois M., Canadell J.G., Dlugokencky E.J., Bergamaschi P., Bergmann D., Blake D.R., Bruhwiler L., et al. Three decades of global methane sources and sinks. Nat. Geosci. 2013;6:813–823. doi: 10.1038/ngeo1955. - DOI
-
- Zinder S.H. Physiological Ecology of Methanogens. In: Ferry J.G., editor. Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. Chapman & Hall; New York, NY, USA: 1993. - DOI
-
- Parkes R.J., Cragg B., Roussel E., Webster G., Weightman A., Sass H. A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere: Geosphere interactions. Marine Geol. 2014;352:409–425. doi: 10.1016/j.margeo.2014.02.009. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

