Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes
- PMID: 33374418
- PMCID: PMC7795494
- DOI: 10.3390/ijms22010125
Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes
Abstract
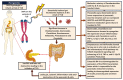
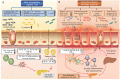
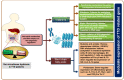
Type 1 diabetes (T1D) is an auto-immune disorder characterized by a complex interaction between the host immune system and various environmental factors in genetically susceptible individuals. Genome-wide association studies (GWAS) identified different T1D risk and protection alleles, however, little is known about the environmental factors that can be linked to these alleles. Recent evidence indicated that, among those environmental factors, dysbiosis (imbalance) in the gut microbiota may play a role in the pathogenesis of T1D, affecting the integrity of the gut and leading to systemic inflammation and auto-destruction of the pancreatic β cells. Several studies have identified changes in the gut microbiome composition in humans and animal models comparing T1D subjects with controls. Those changes were characterized by a higher abundance of Bacteroides and a lower abundance of the butyrate-producing bacteria such as Clostridium clusters IV and XIVa. The mechanisms by which the dysbiotic bacteria and/or their metabolites interact with the genome and/or the epigenome of the host leading to destructive autoimmunity is still not clear. As T1D is a multifactorial disease, understanding the interaction between different environmental factors such as the gut microbiome, the genetic and the epigenetic determinants that are linked with the early appearance of autoantibodies can expand our knowledge about the disease pathogenesis. This review aims to provide insights into the interaction between the gut microbiome, susceptibility genes, epigenetic factors, and the immune system in the pathogenesis of T1D.
Keywords: HLA; NOD mice; immuno-regulation; intestinal permeability; microbial dysbiosis; short-chain fatty acid; single nucleotide polymorphism; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kemppainen K.M., Ardissone A.N., Davis-Richardson A.G., Fagen J.R., Gano K.A., León-Novelo L.G., Vehik K., Casella G., Simell O., Ziegler A.G., et al. Early childhood gut microbiomes show strong geographic differences among subjects at high risk for type 1 diabetes. Diabetes Care. 2015;38:329–332. doi: 10.2337/dc14-0850. - DOI - PMC - PubMed
-
- Blohmé G., Nyström L., Arnqvist H., Lithner F., Littorin B., Olsson P.O., Scherstén B., Wibell L., Ostman J. Male predominance of type 1 (insulin-dependent) diabetes mellitus in young adults: Results from a 5-year prospective nationwide study of the 15–34-year age group in Sweden. Diabetologia. 1992;35:56–62. doi: 10.1007/BF00400852. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

