The Many Faces of CD4+ T Cells: Immunological and Structural Characteristics
- PMID: 33374787
- PMCID: PMC7796221
- DOI: 10.3390/ijms22010073
The Many Faces of CD4+ T Cells: Immunological and Structural Characteristics
Abstract
As a major arm of the cellular immune response, CD4+ T cells are important in the control and clearance of infections. Primarily described as helpers, CD4+ T cells play an integral role in the development and activation of B cells and CD8+ T cells. CD4+ T cells are incredibly heterogeneous, and can be divided into six main lineages based on distinct profiles, namely T helper 1, 2, 17 and 22 (Th1, Th2, Th17, Th22), regulatory T cells (Treg) and T follicular helper cells (Tfh). Recent advances in structural biology have allowed for a detailed characterisation of the molecular mechanisms that drive CD4+ T cell recognition. In this review, we discuss the defining features of the main human CD4+ T cell lineages and their role in immunity, as well as their structural characteristics underlying their detection of pathogens.
Keywords: CD4 T cell; Tfh; Th1; Th17; Th2; Th22; Treg; human leukocyte antigen (HLA); structural biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


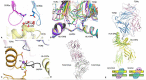
References
-
- Ohno Y., Kitamura H., Takahashi N., Ohtake J., Kaneumi S., Sumida K., Homma S., Kawamura H., Minagawa N., Shibasaki S., et al. IL-6 down-regulates HLA class II expression and IL-12 production of human dendritic cells to impair activation of antigen-specific CD4(+) T cells. Cancer Immunol. Immunother. 2016;65:193–204. doi: 10.1007/s00262-015-1791-4. - DOI - PMC - PubMed
-
- Rijvers L., Melief M.-J., Van Langelaar J., Vries R.M.V.D.V.D., Wierenga-Wolf A.F., Koetzier S.C., Priatel J., Jorritsma T., Van Ham S.M., Hintzen R.Q., et al. The Role of Autoimmunity-Related Gene CLEC16A in the B Cell Receptor-Mediated HLA Class II Pathway. J. Immunol. 2020;205:945–956. doi: 10.4049/jimmunol.1901409. - DOI - PubMed
-
- Codolo G., Toffoletto M., Chemello F., Coletta S., Teixidor G.S., Battaggia G., Munari G., Fassan M., Cagnin S., De Bernard M. Helicobacter pylori Dampens HLA-II Expression on Macrophages via the Up-Regulation of miRNAs Targeting CIITA. Front. Immunol. 2020;10:2923. doi: 10.3389/fimmu.2019.02923. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

