Delivery of Protein Kinase A by CRISPRMAX and Its Effects on Breast Cancer Stem-Like Properties
- PMID: 33374889
- PMCID: PMC7824330
- DOI: 10.3390/pharmaceutics13010011
Delivery of Protein Kinase A by CRISPRMAX and Its Effects on Breast Cancer Stem-Like Properties
Abstract
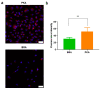
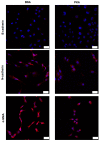
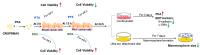
Protein kinase A (PKA) activation has recently been reported to inhibit epithelial-mesenchymal transition (EMT) and cancer stem cell (CSC) ability, which is considered to be responsible for chemoresistance and tumor recurrence in patients. While current studies mainly focus on gene manipulation of the EMT process, the direct delivery of PKA enzymes to cancer cells has never been investigated. Here, we utilize the commercial Lipofectamine CRISPRMAX reagent to directly deliver PKAs to breast cancer cells and evaluate its effects on EMT regulation. We optimized the delivery parameters with fluorescent-labeled bovine serum albumin, and successfully delivered fluorescent PKAs through CRISPRMAX into breast cancer cells. Then, we evaluated the biological effects by immunofluorescence, flow cytometry, mammosphere assay, and chemoresistance assay. Our data showed the expression of EMT-related markers, α-smooth muscle actin and N-cadherin, was downregulated after CRISPRMAX-PKA treatment. Although the CD44+/CD24- population did not change considerably, the size of mammospheres significantly decreased. In paclitaxel and doxorubicin chemoresistance assays, we noticed PKA delivery significantly inhibited paclitaxel resistance rather than doxorubicin resistance. Taken together, these results suggest our direct enzyme delivery can be a potential strategy for inhibiting EMT/CSC-associated traits, providing a safer approach and having more clinical translational efficacy than gene manipulation. This strategy will also facilitate the direct testing of other target enzymes/proteins on their biological functions.
Keywords: CRISPRMAX; cancer stem cells; chemoresistance; drug delivery; epithelial-mesenchymal transition; protein kinase A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

