Molecular and phenotypic diversity of CBL-mutated juvenile myelomonocytic leukemia
- PMID: 33375775
- PMCID: PMC8719097
- DOI: 10.3324/haematol.2020.270595
Molecular and phenotypic diversity of CBL-mutated juvenile myelomonocytic leukemia
Abstract
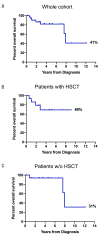
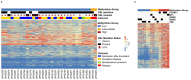
Mutations in the gene CBL were first identified in adults with various myeloid malignancies. Some patients with juvenile myelomonocytic leukemia (JMML) were also noted to harbor mutations in CBL, but were found to have generally less aggressive disease courses compared to other forms of Ras pathway-mutant JMML. Importantly, and in contrast to most reports in adults, the majority of CBL mutations in JMML patients are germline with acquired uniparental disomy occurring in affected marrow cells. Here, we systematically studied a large cohort of 33 JMML patients with CBL mutations and found this disease to be highly diverse in presentation and overall outcome. Moreover, we discovered somatically-acquired CBL mutations in 15% of pediatric patients who presented with more aggressive disease. Neither clinical features nor methylation profiling were able to distinguish somatic CBL patients from germline CBL patients, highlighting the need for germline testing. Overall, we demonstrate that disease courses are quite heterogeneous even among germline CBL patients. Prospective clinical trials are warranted to find ideal treatment strategies for this diverse cohort of patients.
Figures


References
-
- Caye A, Strullu M, Guidez F, et al. . Juvenile myelomonocytic leukemia displays mutations in components of the RAS pathway and the PRC2 network. Nat Genet. 2015;47(11):1334-1340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

