Tissue-selective alternate promoters guide NLRP6 expression
- PMID: 33376129
- PMCID: PMC7772780
- DOI: 10.26508/lsa.202000897
Tissue-selective alternate promoters guide NLRP6 expression
Abstract
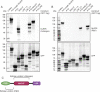
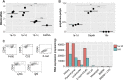
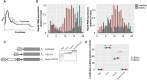
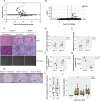
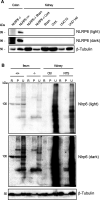
The pryin domain (PYD) domain is involved in protein interactions that lead to assembly of immune-sensing complexes such as inflammasomes. The repertoire of PYD-containing genes expressed by a cell type arms tissues with responses against a range of stimuli. The transcriptional regulation of the PYD gene family however is incompletely understood. Alternative promoter utilization was identified as a mechanism regulating the tissue distribution of human PYD gene family members, including NLRP6 that is translationally silenced outside of intestinal tissue. Results show that alternative transcriptional promoters mediate NLRP6 silencing in mice and humans, despite no upstream genomic synteny. Human NLRP6 contains an internal alternative promoter within exon 2 of the PYD, resulting in a truncated mRNA in nonintestinal tissue. In mice, a proximal promoter was used that expanded the 5' leader sequence restricting nuclear export and abolishing translational efficiency. Nlrp6 was dispensable in disease models targeting the kidney, which expresses noncanonical isoforms. Thus, alternative promoter use is a critical mechanism not just for isoform modulation but for determining expression profile and function of PYD family members.
© 2020 Bracey et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
