IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues
- PMID: 33376221
- PMCID: PMC7776876
- DOI: 10.1073/pnas.2018488117
IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues
Abstract
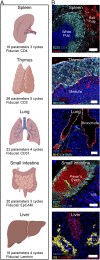
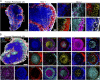
The diverse composition of mammalian tissues poses challenges for understanding the cell-cell interactions required for organ homeostasis and how spatial relationships are perturbed during disease. Existing methods such as single-cell genomics, lacking a spatial context, and traditional immunofluorescence, capturing only two to six molecular features, cannot resolve these issues. Imaging technologies have been developed to address these problems, but each possesses limitations that constrain widespread use. Here we report a method that overcomes major impediments to highly multiplex tissue imaging. "Iterative bleaching extends multiplexity" (IBEX) uses an iterative staining and chemical bleaching method to enable high-resolution imaging of >65 parameters in the same tissue section without physical degradation. IBEX can be employed with various types of conventional microscopes and permits use of both commercially available and user-generated antibodies in an "open" system to allow easy adjustment of staining panels based on ongoing marker discovery efforts. We show how IBEX can also be used with amplified staining methods for imaging strongly fixed tissues with limited epitope retention and with oligonucleotide-based staining, allowing potential cross-referencing between flow cytometry, cellular indexing of transcriptomes and epitopes by sequencing, and IBEX analysis of the same tissue. To facilitate data processing, we provide an open-source platform for automated registration of iterative images. IBEX thus represents a technology that can be rapidly integrated into most current laboratory workflows to achieve high-content imaging to reveal the complex cellular landscape of diverse organs and tissues.
Keywords: high-dimensional imaging; immune system; immunofluorescence; quantitative microscopy; tissue immunity.
Conflict of interest statement
Competing interest statement: J.C. is an employee of BioLegend, which manufactures and sells TotalSeq-A antibodies that are used in one of the workflows described in this paper.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

