Cell fate clusters in ICM organoids arise from cell fate heredity and division: a modelling approach
- PMID: 33376253
- PMCID: PMC7772343
- DOI: 10.1038/s41598-020-80141-3
Cell fate clusters in ICM organoids arise from cell fate heredity and division: a modelling approach
Abstract
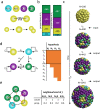
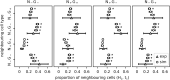
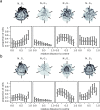
During the mammalian preimplantation phase, cells undergo two subsequent cell fate decisions. During the first decision, the trophectoderm and the inner cell mass are formed. Subsequently, the inner cell mass segregates into the epiblast and the primitive endoderm. Inner cell mass organoids represent an experimental model system, mimicking the second cell fate decision. It has been shown that cells of the same fate tend to cluster stronger than expected for random cell fate decisions. Three major processes are hypothesised to contribute to the cell fate arrangements: (1) chemical signalling; (2) cell sorting; and (3) cell proliferation. In order to quantify the influence of cell proliferation on the observed cell lineage type clustering, we developed an agent-based model accounting for mechanical cell-cell interaction, i.e. adhesion and repulsion, cell division, stochastic cell fate decision and cell fate heredity. The model supports the hypothesis that initial cell fate acquisition is a stochastically driven process, taking place in the early development of inner cell mass organoids. Further, we show that the observed neighbourhood structures can emerge solely due to cell fate heredity during cell division.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. - PubMed
-
- Tarkowski AK, Wróblewska J. Development of blastomeres of mouse eggs isolated at the 4-and 8-cell stage. Development. 1967;18:155–180. - PubMed
-
- Johnson, M. H. & McConnell, J. M. Lineage allocation and cell polarity during mouse embryogenesis. In Seminars in Cell & Developmental Biology, vol. 15, 583–597 (Elsevier, Amsterdam, 2004). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

