Origin and evolution of a gibberellin-deactivating enzyme GAMT
- PMID: 33376939
- PMCID: PMC7762392
- DOI: 10.1002/pld3.287
Origin and evolution of a gibberellin-deactivating enzyme GAMT
Abstract
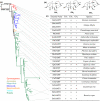
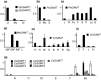
Gibberellins (GAs) are a major class of plant hormones that regulates diverse developmental programs. Both acquiring abilities to synthesize GAs and evolving divergent GA receptors have been demonstrated to play critical roles in the evolution of land plants. In contrast, little is understood regarding the role of GA-inactivating mechanisms in plant evolution. Here we report on the origin and evolution of GA methyltransferases (GAMTs), enzymes that deactivate GAs by converting bioactive GAs to inactive GA methylesters. Prior to this study, GAMT genes, which belong to the SABATH family, were known only from Arabidopsis. Through systematic searches for SABATH genes in the genomes of 260 sequenced land plants and phylogenetic analyses, we have identified a putative GAMT clade specific to seed plants. We have further demonstrated that both gymnosperm and angiosperm representatives of this clade encode active methyltransferases for GA methylation, indicating that they are functional orthologs of GAMT. In seven selected seed plants, GAMT genes were mainly expressed in flowers and/or seeds, indicating a conserved biological role in reproduction. GAMT genes are represented by a single copy in most species, if present, but multiple copies mainly produced by whole genome duplications have been retained in Brassicaceae. Surprisingly, more than 2/3 of the 248 flowering plants examined here lack GAMT genes, including all species of Poales (e.g., grasses), Fabales (legumes), and the large Superasterid clade of eudicots. With these observations, we discuss the significance of GAMT origination, functional conservation and diversification, and frequent loss during the evolution of flowering plants.
Keywords: gene loss; plant hormone; reproduction; seed plants.
© 2020 The Authors. Plant Direct published by American Society of Plant Biologists, Society for Experimental Biology and John Wiley & Sons Ltd.
Figures



References
-
- Chaiprasongsuk, M. , Zhang, C. , Qian, P. , Chen, X. , Li, G. , Trigiano, R. N. , Guo, H. , & Chen, F. (2018). Biochemical characterization in Norway spruce (Picea abies) of SABATH methyltransferases that methylate phytohormones. Phytochemistry, 149, 146–154. 10.1016/j.phytochem.2018.02.010 - DOI - PubMed
LinkOut - more resources
Full Text Sources

