CRISPR-Cas9-Mediated Genomic Deletions Protocol in Zebrafish
- PMID: 33377102
- PMCID: PMC7757680
- DOI: 10.1016/j.xpro.2020.100208
CRISPR-Cas9-Mediated Genomic Deletions Protocol in Zebrafish
Abstract
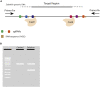
Since its first application for site-directed mutagenesis, the CRISPR-Cas9 system has revolutionized genome engineering. Here, we present a validated workflow for the generation of targeted genomic deletions in zebrafish, including the design, cloning, and synthesis of single-guide RNAs and Cas9 mRNA, followed by microinjection in zebrafish embryos and subsequent genotype screening for the establishment of a mutant line. The versatility and efficiency of this pipeline makes the generation of zebrafish models a widely used approach in functional genetics. For complete details on the use and execution of this protocol, please refer to Amorim et al. (2020).
Keywords: CRISPR; Genetics; Genomics; Model Organisms.
© 2020 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Generation of zebrafish models by CRISPR /Cas9 genome editing.Methods Mol Biol. 2015;1254:341-50. doi: 10.1007/978-1-4939-2152-2_24. Methods Mol Biol. 2015. PMID: 25431076
-
Cas9-based genome editing in zebrafish.Methods Enzymol. 2014;546:377-413. doi: 10.1016/B978-0-12-801185-0.00018-0. Methods Enzymol. 2014. PMID: 25398350 Review.
-
Using local chromatin structure to improve CRISPR/Cas9 efficiency in zebrafish.PLoS One. 2017 Aug 11;12(8):e0182528. doi: 10.1371/journal.pone.0182528. eCollection 2017. PLoS One. 2017. PMID: 28800611 Free PMC article.
-
An improved method for precise genome editing in zebrafish using CRISPR-Cas9 technique.Mol Biol Rep. 2021 Feb;48(2):1951-1957. doi: 10.1007/s11033-020-06125-8. Epub 2021 Jan 22. Mol Biol Rep. 2021. PMID: 33481178 Free PMC article.
-
Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish.Methods. 2017 May 15;121-122:77-85. doi: 10.1016/j.ymeth.2017.03.005. Epub 2017 Mar 12. Methods. 2017. PMID: 28300641 Review.
References
-
- Butkus V., Bitinait J., Janulaitis A. A new restriction endonuclease Eco31l recognizing a non-palindromic sequence. Biochim. Biophys. Acta. 1985;826:208–212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

