MiRNA-516a promotes bladder cancer metastasis by inhibiting MMP9 protein degradation via the AKT/FOXO3A/SMURF1 axis
- PMID: 33377649
- PMCID: PMC7752166
- DOI: 10.1002/ctm2.263
MiRNA-516a promotes bladder cancer metastasis by inhibiting MMP9 protein degradation via the AKT/FOXO3A/SMURF1 axis
Abstract
Background: Metastasis is the leading cause of death in patients with bladder cancer (BC). However, current available treatments exert little effects on metastatic BC. Moreover, traditional grading and staging have only a limited ability to identify metastatic BC. Accumulating evidence indicates that the aberrant expression of microRNA is intimately associated with tumor progression. So far, many miRNAs have been identified as molecular targets for cancer diagnosis and therapy. This study focused on the role of miR-516a-5p (miR-516a) in BC.
Methods: MiR-516a expression and its downstream signaling pathway were detected using molecular cell biology and biochemistry approaches and techniques. Fresh clinical BC tissue was used to study the clinicopathological characteristics of patients with different miR-516a expression. The biological functions of miR-516a in BC were tested both in vivo and in vitro.
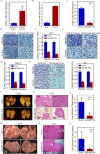
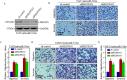
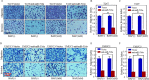
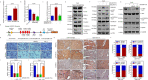
Results: A more invasive BC phenotype was significantly and positively correlated with miR-516a overexpression in BC patients. MiR-516a inhibition significantly decreased BC cell invasion and migration in vitro and in vivo. Furthermore, miR-516a attenuated the expression of PH domain leucine-rich repeat-containing protein phosphatase 2 protein and inhibited SMAD-specific E3 ubiquitin protein ligase 1 transcription by activating the AKT/Forkhead box O3 signaling pathway, which stabilized MMP9 and slowed down its proteasomal degradation, ultimately promoting BC motility and invasiveness.
Conclusions: Our findings reveal the crucial function of miR-516a in promoting BC metastasis, and elucidate the molecular mechanism involved, suggesting that miR-516a may be a promising novel diagnostic and therapeutic target for BC.
Keywords: PHLPP2; bladder cancer; metastasis; miR-516a; migration and invasion.
© 2020 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7‐34. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7‐30. - PubMed
-
- Miller KD, Nogueira L, Mariotto AB, Rowland JH, Siegel RL. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363‐385. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
