The development of neutralizing antibodies against SARS-CoV-2 and their common features
- PMID: 33377928
- PMCID: PMC7799018
- DOI: 10.1093/jmcb/mjaa070
The development of neutralizing antibodies against SARS-CoV-2 and their common features
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a worldwide severe coronavirus disease 2019 (COVID-19) pandemic since December 2019. There is a great demand for effective therapies for the prevention and treatment of COVID-19. Developing therapeutic neutralizing antibodies (NAbs), which could block viral infection, is such a promising approach, as NAbs have been successfully applied to the treatment of other viral infections. The recent advances of antibody technology have greatly accelerated the discovery of SARS-CoV-2 NAbs, and many of which are now actively tested in clinical trials. Here, we review the approaches applied for SARS-CoV-2 NAb development, and discuss the emerging technologies underlining the antibody discovery. We further summarize the common features of these antibodies including the shared neutralizing epitopes and sequence features.
Keywords: COVID-19; RBD; SARS-CoV-2; epitope; neutralizing antibody.
© The Author(s) (2020). Published by Oxford University Press on behalf of Journal of Molecular Cell Biology, IBCB, SIBS, CAS.
Figures
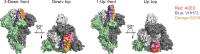
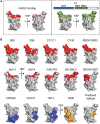

References
-
- Alt F.W., Blackwell T.K., Yancopoulos G.D. (1985). Immunoglobulin genes in transgenic mice. Trends Genet. 1, 231–236.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

