FLIMJ: An open-source ImageJ toolkit for fluorescence lifetime image data analysis
- PMID: 33378370
- PMCID: PMC7773231
- DOI: 10.1371/journal.pone.0238327
FLIMJ: An open-source ImageJ toolkit for fluorescence lifetime image data analysis
Abstract
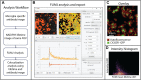
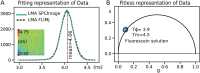
In the field of fluorescence microscopy, there is continued demand for dynamic technologies that can exploit the complete information from every pixel of an image. One imaging technique with proven ability for yielding additional information from fluorescence imaging is Fluorescence Lifetime Imaging Microscopy (FLIM). FLIM allows for the measurement of how long a fluorophore stays in an excited energy state, and this measurement is affected by changes in its chemical microenvironment, such as proximity to other fluorophores, pH, and hydrophobic regions. This ability to provide information about the microenvironment has made FLIM a powerful tool for cellular imaging studies ranging from metabolic measurement to measuring distances between proteins. The increased use of FLIM has necessitated the development of computational tools for integrating FLIM analysis with image and data processing. To address this need, we have created FLIMJ, an ImageJ plugin and toolkit that allows for easy use and development of extensible image analysis workflows with FLIM data. Built on the FLIMLib decay curve fitting library and the ImageJ Ops framework, FLIMJ offers FLIM fitting routines with seamless integration with many other ImageJ components, and the ability to be extended to create complex FLIM analysis workflows. Building on ImageJ Ops also enables FLIMJ's routines to be used with Jupyter notebooks and integrate naturally with science-friendly programming in, e.g., Python and Groovy. We show the extensibility of FLIMJ in two analysis scenarios: lifetime-based image segmentation and image colocalization. We also validate the fitting routines by comparing them against industry FLIM analysis standards.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Rueden CT, Conklin MW, Provenzano PP, Keely PJ, Eliceiri KW. Nonlinear optical microscopy and computational analysis of intrinsic signatures in breast cancer. 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2009. pp. 4077–4080. 10.1109/IEMBS.2009.5334523 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

