Inducible Stem-Cell-Derived Embryos Capture Mouse Morphogenetic Events In Vitro
- PMID: 33378662
- PMCID: PMC7883308
- DOI: 10.1016/j.devcel.2020.12.004
Inducible Stem-Cell-Derived Embryos Capture Mouse Morphogenetic Events In Vitro
Abstract
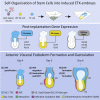
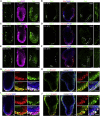
The development of mouse embryos can be partially recapitulated by combining embryonic stem cells (ESCs), trophoblast stem cells (TS), and extra-embryonic endoderm (XEN) stem cells to generate embryo-like structures called ETX embryos. Although ETX embryos transcriptionally capture the mouse gastrula, their ability to recapitulate complex morphogenic events such as gastrulation is limited, possibly due to the limited potential of XEN cells. To address this, we generated ESCs transiently expressing transcription factor Gata4, which drives the extra-embryonic endoderm fate, and combined them with ESCs and TS cells to generate induced ETX embryos (iETX embryos). We show that iETX embryos establish a robust anterior signaling center that migrates unilaterally to break embryo symmetry. Furthermore, iETX embryos gastrulate generating embryonic and extra-embryonic mesoderm and definitive endoderm. Our findings reveal that replacement of XEN cells with ESCs transiently expressing Gata4 endows iETX embryos with greater developmental potential, thus enabling the study of the establishment of anterior-posterior patterning and gastrulation in an in vitro system.
Keywords: EMT; embryogenesis; gastrulation; gastruloid; induced ETX-embryos; stem cells.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Arnold S.J., Robertson E.J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009;10:91–103. - PubMed
-
- Bedzhov I., Leung C.Y., Bialecka M., Zernicka-Goetz M. In vitro culture of mouse blastocysts beyond the implantation stages. Nat. Protoc. 2014;9:2732–2739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

