DNA and RNA Cleavage Complexes and Repair Pathway for TOP3B RNA- and DNA-Protein Crosslinks
- PMID: 33378676
- PMCID: PMC7859927
- DOI: 10.1016/j.celrep.2020.108569
DNA and RNA Cleavage Complexes and Repair Pathway for TOP3B RNA- and DNA-Protein Crosslinks
Abstract
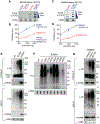
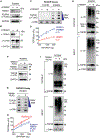
The present study demonstrates that topoisomerase 3B (TOP3B) forms both RNA and DNA cleavage complexes (TOP3Bccs) in vivo and reveals a pathway for repairing TOP3Bccs. For inducing and detecting cellular TOP3Bccs, we engineer a "self-trapping" mutant of TOP3B (R338W-TOP3B). Transfection with R338W-TOP3B induces R-loops, genomic damage, and growth defect, which highlights the importance of TOP3Bcc repair mechanisms. To determine how cells repair TOP3Bccs, we deplete tyrosyl-DNA phosphodiesterases (TDP1 and TDP2). TDP2-deficient cells show elevated TOP3Bccs both in DNA and RNA. Conversely, overexpression of TDP2 lowers cellular TOP3Bccs. Using recombinant human TDP2, we demonstrate that TDP2 can process both denatured and proteolyzed TOP3Bccs. We also show that cellular TOP3Bccs are ubiquitinated by the E3 ligase TRIM41 before undergoing proteasomal processing and excision by TDP2.
Keywords: DNA repair; TDP2; TRIM41; topoisomerase; ubiquitin-proteasome.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures





References
-
- Castillo-Guzman D, Hartono SR, Sanz LA, and Chédin F (2020). SF3B1-targeted Splicing Inhibition Triggers Global Alterations in Transcriptional Dynamics and R-Loop Metabolism. bioRxiv, 2020.2006.2008.130583.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

