Genomic Sequencing and Comparison of Sacbrood Viruses from Apis cerana and Apis mellifera in Taiwan
- PMID: 33379158
- PMCID: PMC7824188
- DOI: 10.3390/pathogens10010014
Genomic Sequencing and Comparison of Sacbrood Viruses from Apis cerana and Apis mellifera in Taiwan
Abstract
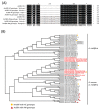
Sacbrood virus (SBV) was the first identified bee virus and shown to cause serious epizootic infections in the population of Apis cerana in Taiwan in 2015. Herein, the whole genome sequences of SBVs in A. cerana and A. mellifera were decoded and designated AcSBV-TW and AmSBV-TW, respectively. The whole genomes of AcSBV-TW and AmSBV-TW were 8776 and 8885 bp, respectively, and shared 90% identity. Each viral genome encoded a polyprotein, which consisted of 2841 aa in AcSBV-TW and 2859 aa in AmSBV-TW, and these sequences shared 95% identity. Compared to 54 other SBVs, the structural protein and protease regions showed high variation, while the helicase was the most highly conserved region among SBVs. Moreover, a 17-amino-acid deletion was found in viral protein 1 (VP1) region of AcSBV-TW compared to AmSBV-TW. The phylogenetic analysis based on the polyprotein sequences and partial VP1 region indicated that AcSBV-TW was grouped into the SBV clade with the AC-genotype (17-aa deletion) and was closely related to AmSBV-SDLY and CSBV-FZ, while AmSBV-TW was grouped into the AM-genotype clade but branched independently from other AmSBVs, indicating that the divergent genomic characteristics of AmSBV-TW might be a consequence of geographic distance driving evolution, and AcSBV-TW was closely related to CSBV-FZ, which originated from China. This 17-amino-acid deletion could be found in either AcSBV or AmSBV in Taiwan, indicating cross-infection between the two viruses. Our data revealed geographic and host specificities between SBVs. The amino acid difference in the VP1 region might serve as a molecular marker for describing SBV cross-infection.
Keywords: Apis cerana; Apis mellifera; sacbrood disease; sacbrood virus.
Conflict of interest statement
The authors declare there is no conflict of interest involved in this work.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

