Identification of New Helicobacter pylori Subpopulations in Native Americans and Mestizos From Peru
- PMID: 33381095
- PMCID: PMC7767971
- DOI: 10.3389/fmicb.2020.601839
Identification of New Helicobacter pylori Subpopulations in Native Americans and Mestizos From Peru
Abstract
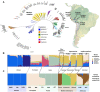
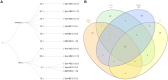
Region-specific Helicobacter pylori subpopulations have been identified. It is proposed that the hspAmerind subpopulation is being displaced from the Americans by an hpEurope population following the conquest. Our study aimed to describe the genomes and methylomes of H. pylori isolates from distinct Peruvian communities: 23 strains collected from three groups of Native Americans (Asháninkas [ASHA, n = 9], Shimaas [SHIM, n = 5] from Amazonas, and Punos from the Andean highlands [PUNO, n = 9]) and 9 modern mestizos from Lima (LIM). Closed genomes and DNA modification calls were obtained using SMRT/PacBio sequencing. We performed evolutionary analyses and evaluated genomic/epigenomic differences among strain groups. We also evaluated human genome-wide data from 74 individuals from the selected Native communities (including the 23 H. pylori strains donors) to compare host and bacterial backgrounds. There were varying degrees of hspAmerind ancestry in all strains, ranging from 7% in LIM to 99% in SHIM. We identified three H. pylori subpopulations corresponding to each of the Native groups and a novel hspEuropePeru which evolved in the modern mestizos. The divergence of the indigenous H. pylori strains recapitulated the genetic structure of Native Americans. Phylogenetic profiling showed that Orthogroups in the indigenous strains seem to have evolved differentially toward epigenomic regulation and chromosome maintenance, whereas OGs in the modern mestizo (LIM) seem to have evolved toward virulence and adherence. The prevalence of cagA +/vacA s1i1m1 genotype was similar across populations (p = 0.32): 89% in ASHA, 67% in PUNO, 56% in LIM and 40% in SHIM. Both cagA and vacA sequences showed that LIM strains were genetically differentiated (p < 0.001) as compared to indigenous strains. We identified 642 R-M systems with 39% of the associated genes located in the core genome. We found 692 methylation motifs, including 254 population-specific sequences not previously described. In Peru, hspAmerind is not extinct, with traces found even in a heavily admixed mestizo population. Notably, our study identified three new hspAmerind subpopulations, one per Native group; and a new subpopulation among mestizos that we named hspEuropePeru. This subpopulation seems to have more virulence-related elements than hspAmerind. Purifying selection driven by variable host immune response may have shaped the evolution of Peruvian subpopulations, potentially impacting disease outcomes.
Keywords: Amerindians; Peru; ancestry; hspAmerind; indigenous; mestizo.
Copyright © 2020 Gutiérrez-Escobar, Velapatiño, Borda, Rabkin, Tarazona-Santos, Cabrera, Cok, Hooper, Jahuira-Arias, Herrera, Noureen, Wang, Romero-Gallo, Tran, Peek, Berg, Gilman and Camargo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Borda V., Alvim I., Aquino M. M., Silva C., Soares-Souza G. B., Leal T. P., et al. (2020). The genetic structure and adaptation of Andean highlanders and Amazonian dwellers is influenced by the interplay between geography and culture. bioRxiv [Preprint] 10.1101/2020.01.30.916270 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

