CRAGE-mediated insertion of fluorescent chromosomal markers for accurate and scalable measurement of co-culture dynamics in Escherichia coli
- PMID: 33381654
- PMCID: PMC7751189
- DOI: 10.1093/synbio/ysaa015
CRAGE-mediated insertion of fluorescent chromosomal markers for accurate and scalable measurement of co-culture dynamics in Escherichia coli
Abstract
Monitoring population dynamics in co-culture is necessary in engineering microbial consortia involved in distributed metabolic processes or biosensing applications. However, it remains difficult to measure strain-specific growth dynamics in high-throughput formats. This is especially vexing in plate-based functional screens leveraging whole-cell biosensors to detect specific metabolic signals. Here, we develop an experimental high-throughput co-culture system to measure and model the relationship between fluorescence and cell abundance, combining chassis-independent recombinase-assisted genome engineering (CRAGE) and whole-cell biosensing with a PemrR-green fluorescent protein (GFP) monoaromatic reporter used in plate-based functional screening. CRAGE was used to construct Escherichia coli EPI300 strains constitutively expressing red fluorescent protein (RFP) and the relationship between RFP expression and optical density (OD600) was determined throughout the EPI300 growth cycle. A linear equation describing the increase of normalized RFP fluorescence during deceleration phase was derived and used to predict biosensor strain dynamics in co-culture. Measured and predicted values were compared using flow cytometric detection methods. Induction of the biosensor lead to increased GFP fluorescence normalized to biosensor cell abundance, as expected, but a significant decrease in relative abundance of the biosensor strain in co-culture and a decrease in bulk GFP fluorescence. Taken together, these results highlight sensitivity of population dynamics to variations in metabolic activity in co-culture and the potential effect of these dynamics on the performance of functional screens in plate-based formats. The engineered strains and model used to evaluate these dynamics provide a framework for optimizing growth of synthetic co-cultures used in screening, testing and pathway engineering applications.
Keywords: CRAGE; biosensors; co-culture dynamics; functional screening; microbial consortia.
© The Author(s) 2020. Published by Oxford University Press.
Figures






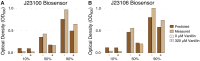
Similar articles
-
Naringenin-responsive riboswitch-based fluorescent biosensor module for Escherichia coli co-cultures.Biotechnol Bioeng. 2017 Oct;114(10):2235-2244. doi: 10.1002/bit.26340. Epub 2017 Jun 5. Biotechnol Bioeng. 2017. PMID: 28543037
-
Engineering tunable biosensors for monitoring putrescine in Escherichia coli.Biotechnol Bioeng. 2018 Apr;115(4):1014-1027. doi: 10.1002/bit.26521. Epub 2018 Jan 8. Biotechnol Bioeng. 2018. PMID: 29251347
-
Reactor control system in bacterial co-culture based on fluorescent proteins using an Arduino-based home-made device.Biotechnol J. 2021 Dec;16(12):e2100169. doi: 10.1002/biot.202100169. Epub 2021 Oct 7. Biotechnol J. 2021. PMID: 34553835
-
Fluorescent biosensors for drug discovery new tools for old targets--screening for inhibitors of cyclin-dependent kinases.Eur J Med Chem. 2014 Dec 17;88:74-88. doi: 10.1016/j.ejmech.2014.10.003. Epub 2014 Oct 5. Eur J Med Chem. 2014. PMID: 25314935 Review.
-
Biosensors design in yeast and applications in metabolic engineering.FEMS Yeast Res. 2019 Dec 1;19(8):foz082. doi: 10.1093/femsyr/foz082. FEMS Yeast Res. 2019. PMID: 31778177 Review.
Cited by
-
Expression of Fluorescence Reporters and Natural Products in Native Gut Escherichia coli.ACS Synth Biol. 2025 May 16;14(5):1557-1566. doi: 10.1021/acssynbio.4c00835. Epub 2025 Mar 26. ACS Synth Biol. 2025. PMID: 40138712 Free PMC article.
-
Measuring differential fitness costs and interactions between genetic cassettes using fluorescent spectrophotometry.Appl Environ Microbiol. 2024 Feb 21;90(2):e0141923. doi: 10.1128/aem.01419-23. Epub 2024 Feb 1. Appl Environ Microbiol. 2024. PMID: 38299817 Free PMC article.
-
Measurement Techniques to Resolve and Control Population Dynamics of Mixed-Culture Processes.Trends Biotechnol. 2021 Oct;39(10):1093-1109. doi: 10.1016/j.tibtech.2021.01.006. Epub 2021 Feb 8. Trends Biotechnol. 2021. PMID: 33573846 Free PMC article. Review.
-
Methionine-producing tumor micro(be) environment fuels growth of solid tumors.Cell Oncol (Dordr). 2023 Dec;46(6):1659-1673. doi: 10.1007/s13402-023-00832-7. Epub 2023 Jun 15. Cell Oncol (Dordr). 2023. PMID: 37318751 Free PMC article.
References
-
- Hallam S.J., McCutcheon J.P. (2015) Microbes don't play solitaire: how cooperation trumps isolation in the microbial world. Environ. Microbiol. Rep., 7, 392–398. - PubMed
-
- Hays S.G., Patrick W.G., Ziesack M., Oxman N., Silver P.A. (2015) Better together: engineering and application of microbial symbioses. Curr. Opin. Biotechnol., 36, 40–49. - PubMed
