Using a GTR+Γ substitution model for dating sequence divergence when stationarity and time-reversibility assumptions are violated
- PMID: 33381826
- PMCID: PMC7773479
- DOI: 10.1093/bioinformatics/btaa820
Using a GTR+Γ substitution model for dating sequence divergence when stationarity and time-reversibility assumptions are violated
Abstract
Motivation: As the number and diversity of species and genes grow in contemporary datasets, two common assumptions made in all molecular dating methods, namely the time-reversibility and stationarity of the substitution process, become untenable. No software tools for molecular dating allow researchers to relax these two assumptions in their data analyses. Frequently the same General Time Reversible (GTR) model across lineages along with a gamma (+Γ) distributed rates across sites is used in relaxed clock analyses, which assumes time-reversibility and stationarity of the substitution process. Many reports have quantified the impact of violations of these underlying assumptions on molecular phylogeny, but none have systematically analyzed their impact on divergence time estimates.
Results: We quantified the bias on time estimates that resulted from using the GTR + Γ model for the analysis of computer-simulated nucleotide sequence alignments that were evolved with non-stationary (NS) and non-reversible (NR) substitution models. We tested Bayesian and RelTime approaches that do not require a molecular clock for estimating divergence times. Divergence times obtained using a GTR + Γ model differed only slightly (∼3% on average) from the expected times for NR datasets, but the difference was larger for NS datasets (∼10% on average). The use of only a few calibrations reduced these biases considerably (∼5%). Confidence and credibility intervals from GTR + Γ analysis usually contained correct times. Therefore, the bias introduced by the use of the GTR + Γ model to analyze datasets, in which the time-reversibility and stationarity assumptions are violated, is likely not large and can be reduced by applying multiple calibrations.
Availability and implementation: All datasets are deposited in Figshare: https://doi.org/10.6084/m9.figshare.12594638.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

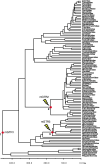




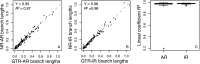
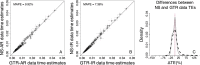







References
-
- Blanquart S., Lartillot N. (2006) A Bayesian compound stochastic process for modeling non-stationary and non-homogeneous sequence evolution. Mol. Biol. Evol., 23, 2058–2071. - PubMed
-
- Blanquart S., Lartillot N. (2008) A site- and time-heterogeneous model of amino acid replacement. Mol. Biol. Evol., 25, 842–858. - PubMed
-
- dos Reis M., Yang Z. (2011) Approximate likelihood calculation on a phylogeny for Bayesian Estimation of Divergence Times. Mol. Biol. Evol., 28, 2161–2172. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

