Feasible-metabolic-pathway-exploration technique using chemical latent space
- PMID: 33381845
- PMCID: PMC8454040
- DOI: 10.1093/bioinformatics/btaa809
Feasible-metabolic-pathway-exploration technique using chemical latent space
Abstract
Motivation: Exploring metabolic pathways is one of the key techniques for developing highly productive microbes for the bioproduction of chemical compounds. To explore feasible pathways, not only examining a combination of well-known enzymatic reactions but also finding potential enzymatic reactions that can catalyze the desired structural changes are necessary. To achieve this, most conventional techniques use manually predefined-reaction rules, however, they cannot sufficiently find potential reactions because the conventional rules cannot comprehensively express structural changes before and after enzymatic reactions. Evaluating the feasibility of the explored pathways is another challenge because there is no way to validate the reaction possibility of unknown enzymatic reactions by these rules. Therefore, a technique for comprehensively capturing the structural changes in enzymatic reactions and a technique for evaluating the pathway feasibility are still necessary to explore feasible metabolic pathways.
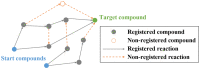
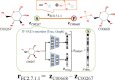
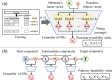
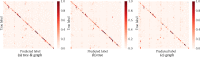
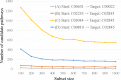
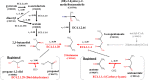
Results: We developed a feasible-pathway-exploration technique using chemical latent space obtained from a deep generative model for compound structures. With this technique, an enzymatic reaction is regarded as a difference vector between the main substrate and the main product in chemical latent space acquired from the generative model. Features of the enzymatic reaction are embedded into the fixed-dimensional vector, and it is possible to express structural changes of enzymatic reactions comprehensively. The technique also involves differential-evolution-based reaction selection to design feasible candidate pathways and pathway scoring using neural-network-based reaction-possibility prediction. The proposed technique was applied to the non-registered pathways relevant to the production of 2-butanone, and successfully explored feasible pathways that include such reactions.
© The Author(s) 2020. Published by Oxford University Press.
Figures





Similar articles
-
Curating a comprehensive set of enzymatic reaction rules for efficient novel biosynthetic pathway design.Metab Eng. 2021 May;65:79-87. doi: 10.1016/j.ymben.2021.02.006. Epub 2021 Mar 1. Metab Eng. 2021. PMID: 33662575
-
Supervised de novo reconstruction of metabolic pathways from metabolome-scale compound sets.Bioinformatics. 2013 Jul 1;29(13):i135-44. doi: 10.1093/bioinformatics/btt244. Bioinformatics. 2013. PMID: 23812977 Free PMC article.
-
An efficient algorithm for de novo predictions of biochemical pathways between chemical compounds.BMC Bioinformatics. 2012;13 Suppl 17(Suppl 17):S8. doi: 10.1186/1471-2105-13-S17-S8. Epub 2012 Dec 13. BMC Bioinformatics. 2012. PMID: 23282285 Free PMC article.
-
Computational tools and resources for designing new pathways to small molecules.Curr Opin Biotechnol. 2022 Aug;76:102722. doi: 10.1016/j.copbio.2022.102722. Epub 2022 Apr 26. Curr Opin Biotechnol. 2022. PMID: 35483185 Review.
-
The widespread role of non-enzymatic reactions in cellular metabolism.Curr Opin Biotechnol. 2015 Aug;34:153-61. doi: 10.1016/j.copbio.2014.12.020. Epub 2015 Jan 22. Curr Opin Biotechnol. 2015. PMID: 25617827 Free PMC article. Review.
Cited by
-
Designing Microbial Cell Factories for the Production of Chemicals.JACS Au. 2022 Aug 4;2(8):1781-1799. doi: 10.1021/jacsau.2c00344. eCollection 2022 Aug 22. JACS Au. 2022. PMID: 36032533 Free PMC article. Review.
References
-
- Araki M. et al. (2014) M-path: a compass for navigating potential metabolic pathways. Bioinformatics, 31, 905–911. - PubMed
-
- Battiti R., Colla A.M. (1994) Democracy in neural nets: voting schemes for classification. Neural Networks, 7, 691–707.
-
- Choi K.R. et al. (2019) Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol., 37, 817–837. - PubMed

