Fisetin 8-C-glucoside as entry inhibitor in SARS CoV-2 infection: molecular modelling study
- PMID: 33382023
- PMCID: PMC7784833
- DOI: 10.1080/07391102.2020.1868335
Fisetin 8-C-glucoside as entry inhibitor in SARS CoV-2 infection: molecular modelling study
Abstract
Coronaviruses are RNA viruses that infect varied species including humans. TMPRSS2 is gateway for SARS CoV-2 entry into the host cell. It causes proteolytic activation of spike protein and discharge of the peptide into host cell. The TMPRSS2 inhibition could be one of the approaches to stop the viral entry, therefore, interaction pattern and binding energies for Fisetin and TMPRSS2 have been explored in the present study. TMPRSS2 peptide was used for homology modelling and then for further study. Molecular docking score and MMGBSA Binding energy of Fisetin was better than Nafamostat, a known inhibitor of TMPRSS2. Post docking MM-GBSA free energy for Fisetin and Nafamostat was -42.78 and -21.11 kcal/mol, respectively. Fisetin forms H bond with Val 25, His 41, Lys 42, Lys 45, Glu 44, Ser186. Nafamostat formed H bonds with Lys 85, Asp 90, Asp 203. RMSD plots of TMPRSS2, TMPRSS2-Fisetin and TMPRSS2-Nafamostat complex showed stable profile with very small fluctuation during entire simulation of 150 ns. Significant decrease in TMPRSS2-Fisetin and TMPRSS2-Nafamostat complex fluctuation occurred around His 41, Glu 44, Gly 136, Ser 186 in RMSF study. During simulation Fisetin interaction was observed with residues Val 25, His 41, Glu 44, Lys 45, Lys 87, Gly 136, Gln 183, Ser 186 likewise interaction of Nafamostat with Lys 85, Asp 90, Asn 163, Asp 203 and Ser 205. Post simulation MM-GBSA free energy was found to be -51.87 ± 4.3 and -48.23 ± 4.39 kcal/mol for TMPRSS2 with Fisetin and Nafamostat, respectively.Communicated by Ramaswamy H. Sarma.
Keywords: Fisetin; MD simulation; MMGBSA; TMPRSS2; homology modelling.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
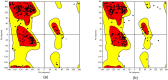



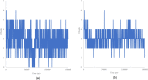




Similar articles
-
Strong Binding of Leupeptin with TMPRSS2 Protease May Be an Alternative to Camostat and Nafamostat for SARS-CoV-2 Repurposed Drug: Evaluation from Molecular Docking and Molecular Dynamics Simulations.Appl Biochem Biotechnol. 2021 Jun;193(6):1909-1923. doi: 10.1007/s12010-020-03475-8. Epub 2021 Jan 29. Appl Biochem Biotechnol. 2021. PMID: 33512650 Free PMC article.
-
Identification, virtual screening and molecular dynamic analysis of novel TMPRSS2 inhibitors from natural compound database as potential entry-blocking agents in SARS-CoV-2 therapy.Struct Chem. 2022;33(5):1609-1617. doi: 10.1007/s11224-022-01991-3. Epub 2022 Jun 21. Struct Chem. 2022. PMID: 35754942 Free PMC article.
-
The primary structure of human liver manganese superoxide dismutase.J Biol Chem. 1984 Oct 25;259(20):12595-601. J Biol Chem. 1984. PMID: 6386798
-
Molecular docking analysis reveals the functional inhibitory effect of Genistein and Quercetin on TMPRSS2: SARS-COV-2 cell entry facilitator spike protein.BMC Bioinformatics. 2022 May 16;23(1):180. doi: 10.1186/s12859-022-04724-9. BMC Bioinformatics. 2022. PMID: 35578172 Free PMC article.
-
Leucoefdin a potential inhibitor against SARS CoV-2 Mpro.J Biomol Struct Dyn. 2021 Aug;39(12):4427-4432. doi: 10.1080/07391102.2020.1777903. Epub 2020 Jun 17. J Biomol Struct Dyn. 2021. PMID: 34281489 Free PMC article.
Cited by
-
Structure-guided discovery of a novel BTK inhibitor inducing apoptosis and G1 phase arrest in tumor cells.Mol Divers. 2025 Aug 31. doi: 10.1007/s11030-025-11334-z. Online ahead of print. Mol Divers. 2025. PMID: 40886252
-
Are Antiviral Flavonoids Part of the Solution to the COVID-19 Pandemic?Integr Med (Encinitas). 2021 Dec;20(6):8-13. Integr Med (Encinitas). 2021. PMID: 35250397 Free PMC article.
-
Discovery of Histone Deacetylase Inhibitor Using Molecular Modeling and Free Energy Calculations.ACS Omega. 2022 May 24;7(22):18786-18794. doi: 10.1021/acsomega.2c01572. eCollection 2022 Jun 7. ACS Omega. 2022. PMID: 35694501 Free PMC article.
References
-
- Alireza, P., Hosseini, M. M., & Akhavan-Niaki, H. (2020). First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. Journal of Biomolecular Structure and Dynamics, 10.1080/07391102.2020.1767690 - DOI - PMC - PubMed
-
- Bertram, S., Heurich, A., Lavender, H., Gierer, S., Danisch, S., Perin, P., Lucas, J. M., Nelson, P. S., Pöhlmann, S., & Soilleux, E. J. (2012). Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS One, 7(4), e35876. 10.1371/journal.pone.0035876 - DOI - PMC - PubMed
-
- Christ, C. D., Mark, A. E., & van Gunsteren, W. F. (2009). Basic ingredients of free energy calculations: A review. Journal of Computational Chemistry, 31, 1569–1582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
