Translation initiation and its relevance in colorectal cancer
- PMID: 33382175
- PMCID: PMC9291299
- DOI: 10.1111/febs.15690
Translation initiation and its relevance in colorectal cancer
Abstract
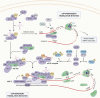
Protein synthesis is one of the most essential processes in every kingdom of life, and its dysregulation is a known driving force in cancer development. Multiple signaling pathways converge on the translation initiation machinery, and this plays a crucial role in regulating differential gene expression. In colorectal cancer, dysregulation of initiation results in translational reprogramming, which promotes the selective translation of mRNAs required for many oncogenic processes. The majority of upstream mutations found in colorectal cancer, including alterations in the WNT, MAPK, and PI3K\AKT pathways, have been demonstrated to play a significant role in translational reprogramming. Many translation initiation factors are also known to be dysregulated, resulting in translational reprogramming during tumor initiation and/or maintenance. In this review, we outline the role of translational reprogramming that occurs during colorectal cancer development and progression and highlight some of the most critical factors affecting the etiology of this disease.
Keywords: RNA translation; colorectal cancer; protein synthesis; translation initiation; translational control.
© 2021 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

