Adaptability of the ubiquitin-proteasome system to proteolytic and folding stressors
- PMID: 33382395
- PMCID: PMC7780722
- DOI: 10.1083/jcb.201912041
Adaptability of the ubiquitin-proteasome system to proteolytic and folding stressors
Abstract
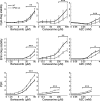
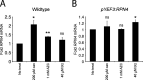
Aging, disease, and environmental stressors are associated with failures in the ubiquitin-proteasome system (UPS), yet a quantitative understanding of how stressors affect the proteome and how the UPS responds is lacking. Here we assessed UPS performance and adaptability in yeast under stressors using quantitative measurements of misfolded substrate stability and stress-dependent UPS regulation by the transcription factor Rpn4. We found that impairing degradation rates (proteolytic stress) and generating misfolded proteins (folding stress) elicited distinct effects on the proteome and on UPS adaptation. Folding stressors stabilized proteins via aggregation rather than overburdening the proteasome, as occurred under proteolytic stress. Still, the UPS productively adapted to both stressors using separate mechanisms: proteolytic stressors caused Rpn4 stabilization while folding stressors increased RPN4 transcription. In some cases, adaptation completely prevented loss of UPS substrate degradation. Our work reveals the distinct effects of proteotoxic stressors and the versatility of cells in adapting the UPS.
© 2020 Work and Brandman.
Figures









Similar articles
-
Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae.PLoS Genet. 2011 Sep;7(9):e1002253. doi: 10.1371/journal.pgen.1002253. Epub 2011 Sep 8. PLoS Genet. 2011. PMID: 21931558 Free PMC article.
-
Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions.Genetics. 2010 Feb;184(2):335-42. doi: 10.1534/genetics.109.112227. Epub 2009 Nov 23. Genetics. 2010. PMID: 19933873 Free PMC article.
-
Distinct classes of misfolded proteins differentially affect the growth of yeast compromised for proteasome function.FEBS Lett. 2021 Sep;595(18):2383-2394. doi: 10.1002/1873-3468.14172. Epub 2021 Aug 17. FEBS Lett. 2021. PMID: 34358326 Free PMC article.
-
Protein Quality Control of the Endoplasmic Reticulum and Ubiquitin-Proteasome-Triggered Degradation of Aberrant Proteins: Yeast Pioneers the Path.Annu Rev Biochem. 2018 Jun 20;87:751-782. doi: 10.1146/annurev-biochem-062917-012749. Epub 2018 Feb 2. Annu Rev Biochem. 2018. PMID: 29394096 Review.
-
We Are What We Eat: Ubiquitin-Proteasome System (UPS) Modulation Through Dietary Products.Adv Exp Med Biol. 2020;1233:329-348. doi: 10.1007/978-3-030-38266-7_15. Adv Exp Med Biol. 2020. PMID: 32274765 Review.
Cited by
-
The Protein Response of Salt-Tolerant Zygosaccharomyces rouxii to High-Temperature Stress during the Lag Phase.J Fungi (Basel). 2024 Jan 5;10(1):48. doi: 10.3390/jof10010048. J Fungi (Basel). 2024. PMID: 38248957 Free PMC article.
-
The Heat Shock Response as a Condensate Cascade.J Mol Biol. 2024 Jul 15;436(14):168642. doi: 10.1016/j.jmb.2024.168642. Epub 2024 Jun 5. J Mol Biol. 2024. PMID: 38848866 Free PMC article. Review.
-
Genotype-by-environment interactions shape ubiquitin-proteasome system activity.bioRxiv [Preprint]. 2024 Nov 21:2024.11.21.624644. doi: 10.1101/2024.11.21.624644. bioRxiv. 2024. PMID: 39605480 Free PMC article. Preprint.
-
26S proteasomes become stably activated upon heat shock when ubiquitination and protein degradation increase.Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2122482119. doi: 10.1073/pnas.2122482119. Epub 2022 Jun 15. Proc Natl Acad Sci U S A. 2022. PMID: 35704754 Free PMC article.
-
The APC/C Activator Cdh1p Plays a Role in Mitochondrial Metabolic Remodelling in Yeast.Int J Mol Sci. 2023 Feb 18;24(4):4111. doi: 10.3390/ijms24044111. Int J Mol Sci. 2023. PMID: 36835555 Free PMC article.
References
-
- Brandman, O., Stewart-Ornstein J., Wong D., Larson A., Williams C.C., Li G.-W., Zhou S., King D., Shen P.S., Weibezahn J., et al. . 2012. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 151:1042–1054. 10.1016/j.cell.2012.10.044 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

