Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism, and accumulation
- PMID: 33382907
- PMCID: PMC8212739
- DOI: 10.1111/cts.12969
Characterizing the reproducibility in using a liver microphysiological system for assaying drug toxicity, metabolism, and accumulation
Abstract
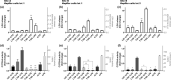
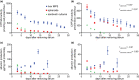
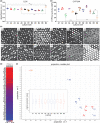
Liver microphysiological systems (MPSs) are promising models for predicting hepatic drug effects. Yet, after a decade since their introduction, MPSs are not routinely used in drug development due to lack of criteria for ensuring reproducibility of results. We characterized the feasibility of a liver MPS to yield reproducible outcomes of experiments assaying drug toxicity, metabolism, and intracellular accumulation. The ability of the liver MPS to reproduce hepatotoxic effects was assessed using trovafloxacin, which increased lactate dehydrogenase (LDH) release and reduced cytochrome P450 3A4 (CYP3A4) activity. These observations were made in two test sites and with different batches of Kupffer cells. Upon culturing equivalent hepatocytes in the MPS, spheroids, and sandwich cultures, differences between culture formats were detected in CYP3A4 activity and albumin production. Cells in all culture formats exhibited different sensitivities to hepatotoxicant exposure. Hepatocytes in the MPS were more functionally stable than those of other culture platforms, as CYP3A4 activity and albumin secretion remained prominent for greater than 18 days in culture, whereas functional decline occurred earlier in spheroids (12 days) and sandwich cultures (7 days). The MPS was also demonstrated to be suitable for metabolism studies, where CYP3A4 activity, troglitazone metabolites, diclofenac clearance, and intracellular accumulation of chloroquine were quantified. To ensure reproducibility between studies with the MPS, the combined use of LDH and CYP3A4 assays were implemented as quality control metrics. Overall results indicated that the liver MPS can be used reproducibly in general drug evaluation applications. Study outcomes led to general considerations and recommendations for using liver MPSs. Study Highlights WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC? Microphysiological systems (MPSs) have been designed to recreate organ- or tissue-specific characteristics of extracellular microenvironments that enhance the physiological relevance of cells in culture. Liver MPSs enable long-lasting and stable culture of hepatic cells by culturing them in three-dimensions and exposing them to fluid flow. WHAT QUESTION DID THIS STUDY ADDRESS? What is the functional performance relative to other cell culture platforms and the reproducibility of a liver MPS for assessing drug development and evaluation questions, such as toxicity, metabolism, and pharmacokinetics? WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE? The liver MPS systematically detected the toxicity of trovafloxacin. When compared with spheroids and sandwich cultures, this system had a more stable function and different sensitivity to troglitazone, tamoxifen, and digoxin. Quantifying phase II metabolism of troglitazone and intracellular accumulation of chloroquine demonstrated the potential use of the liver MPS for studying drug metabolism and pharmacokinetics. Quality control criteria for assessing chip function were key for reliably using the liver MPS. HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE? Due to its functional robustness and physiological relevance (3D culture, cells expose to fluid flow and co-culture of different cell types), the liver MPS can, in a reproducible manner: (i) detect inflammatory-induced drug toxicity, as demonstrated with trovafloxacin, (ii) detect the toxicity of other drugs, such as troglitazone, tamoxifen, and digoxin, with different effects than those detected in spheroids and sandwich cultures, (iii) enable studies of hepatic function that rely on prolonged cellular activity, and (iv) detect phase II metabolites and drug accumulation to potentially support the interpretation of clinical data. The integration of MPSs in drug development will be facilitated by careful evaluation of performance and reproducibility as performed in this study.
© 2020 The Authors. Clinical and Translational Science published by Wiley Periodicals LLC on behalf of the American Society for Clinical Pharmacology and Therapeutics. This article has been contributed to by US Government employees and their work is in the public domain in the USA.
Conflict of interest statement
Dr. Hughes owns stock in and is employed by CN Bio Innovations. Dr. Kostrzewski is employed by CN Bio Innovations. Ms. Miedzik was employed by CN Bio Innovations. All other authors declared no competing interests for this work.
Figures


References
-
- Baudy AR, Otieno MA, Hewitt P, et al. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip. 2020;20:215‐225. - PubMed
-
- Fowler S, Chen WLK, Duignan DB, et al. Microphysiological systems for ADME‐related applications: current status and recommendations for system development and characterization. Lab Chip. 2020;20:446‐467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

