Functional Analysis of Haplotypes in Bovine PSAP Gene and Their Relationship with Beef Cattle Production Traits
- PMID: 33383762
- PMCID: PMC7824473
- DOI: 10.3390/ani11010049
Functional Analysis of Haplotypes in Bovine PSAP Gene and Their Relationship with Beef Cattle Production Traits
Abstract
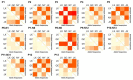
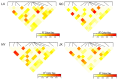
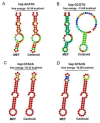
The purpose of this study was to explore functional variants in the prosaposin (PSAP) three prime untranslated region (3' UTR) and clarify the relationship between the variants and morphological traits. Through Sanger sequencing, 13 variations were identified in bovine PSAP in four Chinese cattle breeds, with six of them being loci in 3' UTR. In particular, Nanyang (NY) cattle had a special genotype and haplotype distribution compared to the other three breeds. NY cattle with ACATG and GCGTG haplotypes had higher morphological traits than GTACA and GTACG haplotypes. The results of dual-luciferase reporter assay showed that ACATG and GCGTG haplotypes affected the morphological traits of NY cattle by altering the secondary structure of PSAP 3' UTR rather than the miR-184 target sites. The findings of this study could be an evidence of a complex and varying mechanism between variants and animal morphological traits and could be used to complement candidate genes for molecular breeding.
Keywords: PSAP; bovine; haplotypes; mRNA secondary structure; miR-184; morphological traits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Relationship of polymorphisms within ZBED6 gene and growth traits in beef cattle.Gene. 2013 Sep 10;526(2):107-11. doi: 10.1016/j.gene.2013.04.049. Epub 2013 May 1. Gene. 2013. PMID: 23644023
-
Haplotype combination of SREBP-1c gene sequence variants is associated with growth traits in cattle.Genome. 2011 Jun;54(6):507-16. doi: 10.1139/g11-016. Genome. 2011. PMID: 21639705
-
Exploring of InDel in bovine PSAP gene and their association with growth traits in different development stages.Anim Biotechnol. 2022 Feb;33(1):1-12. doi: 10.1080/10495398.2020.1758122. Epub 2020 May 5. Anim Biotechnol. 2022. PMID: 32367774
-
TNP1 Functional SNPs in bta-miR-532 and bta-miR-204 Target Sites Are Associated with Semen Quality Traits in Chinese Holstein Bulls.Biol Reprod. 2015 Jun;92(6):139. doi: 10.1095/biolreprod.114.126672. Epub 2015 Apr 22. Biol Reprod. 2015. PMID: 25904013
-
Novel polymorphisms of the APOA2 gene and its promoter region affect body traits in cattle.Gene. 2013 Dec 1;531(2):288-93. doi: 10.1016/j.gene.2013.08.081. Epub 2013 Sep 1. Gene. 2013. PMID: 24004543
Cited by
-
Detection of Insertion/Deletions (InDel) Within Five Clock Genes and Their Associations with Growth Traits in Four Chinese Sheep Breeds.Vet Sci. 2025 Jan 9;12(1):39. doi: 10.3390/vetsci12010039. Vet Sci. 2025. PMID: 39852913 Free PMC article.
References
Grants and funding
- 2016CXY-15/the Collaborative Innovation Major Projects of Research and Production Practical Application for Yangling Demonstration Zone
- No. 16190050/the Integrated Demonstration Project of Key Technologies for Cattle and Sheep Breeding and Farming in Farming Pastoral Ecotone of Ministry of Agriculture and Rural Affairs
- CARS-34/China Agriculture Research System
LinkOut - more resources
Full Text Sources
Miscellaneous

