Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells
- PMID: 33383772
- PMCID: PMC7795497
- DOI: 10.3390/ijms22010252
Mitochondrial TSPO Deficiency Triggers Retrograde Signaling in MA-10 Mouse Tumor Leydig Cells
Abstract
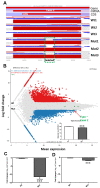
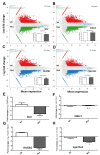
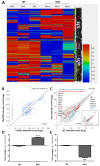
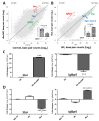
The mitochondrial translocator protein (TSPO) has been shown to bind cholesterol with high affinity and is involved in mediating its availability for steroidogenesis. We recently reported that targeted Tspo gene deletion in MA-10 mouse tumor Leydig cells resulted in reduced cAMP-stimulated steroid formation and significant reduction in the mitochondrial membrane potential (ΔΨm) compared to control cells. We hypothesized that ΔΨm reduction in the absence of TSPO probably reflects the dysregulation and/or maintenance failure of some basic mitochondrial function(s). To explore the consequences of TSPO depletion via CRISPR-Cas9-mediated deletion (indel) mutation in MA-10 cells, we assessed the transcriptome changes in TSPO-mutant versus wild-type (Wt) cells using RNA-seq. Gene expression profiles were validated using real-time PCR. We report herein that there are significant changes in nuclear gene expression in Tspo mutant versus Wt cells. The identified transcriptome changes were mapped to several signaling pathways including the regulation of membrane potential, calcium signaling, extracellular matrix, and phagocytosis. This is a retrograde signaling pathway from the mitochondria to the nucleus and is probably the result of changes in expression of several transcription factors, including key members of the NF-κB pathway. In conclusion, TSPO regulates nuclear gene expression through intracellular signaling. This is the first evidence of a compensatory response to the loss of TSPO with transcriptome changes at the cellular level.
Keywords: RNA-seq; calcium homeostasis; genomic edition; mitochondria; retrograde signaling; translocator protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Papadopoulos V., Baraldi M., Guilarte T.R., Knudsen T.B., Lacapere J.J., Lindemann P., Norenberg M.D., Nutt D., Weizman A., Zhang M.R., et al. Translocator protein (18kDa): New nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol. Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. - DOI - PubMed
-
- Rone M.B., Midzak A.S., Issop L., Rammouz G., Jagannathan S., Fan J., Ye X., Blonder J., Veenstra T., Papadopoulos V. Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol. Endocrinol. 2012;26:1868–1882. doi: 10.1210/me.2012-1159. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

